
🧬Protein Design for CRISPR proteins🧑🏽💻👨🏽🎨
www.biorxiv.org/content/10.1...
1/n

www.biorxiv.org/content/10.1...
1/n

@gocastelobranco.bsky.social lab!! We use MPRA & scCRISPRi/a to interrogate possible functions of GWAS-identified Multiple sclerosis risk SNPs in iPSC-derived oligodendrocytes.
www.biorxiv.org/content/10.1...
@gocastelobranco.bsky.social lab!! We use MPRA & scCRISPRi/a to interrogate possible functions of GWAS-identified Multiple sclerosis risk SNPs in iPSC-derived oligodendrocytes.
www.biorxiv.org/content/10.1...
www.biorxiv.org/cgi/content/...

www.biorxiv.org/content/10.1...


Evolved compact TranC11a matched SpCas9 edit, trait mod in plant/human.




Evolved compact TranC11a matched SpCas9 edit, trait mod in plant/human.
Learn more in the full paper: www.science.org/doi/10.1126/...

Learn more in the full paper: www.science.org/doi/10.1126/...


with generative bioML, i'm always looking at how similar the generated sequences are to known sequences. let's take a look
Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
with generative bioML, i'm always looking at how similar the generated sequences are to known sequences. let's take a look
Our manuscript describes:
1️⃣ Engineering a target-specific BE🧬
2⃣ A *must avoid* bystander edit that occurs with WT SpCas9 BEs! 🙅♂️
3⃣ Extension of lifespan after in vivo editing! 🐁✅
www.nature.com/articles/s41...

Our manuscript describes:
1️⃣ Engineering a target-specific BE🧬
2⃣ A *must avoid* bystander edit that occurs with WT SpCas9 BEs! 🙅♂️
3⃣ Extension of lifespan after in vivo editing! 🐁✅
www.nature.com/articles/s41...


www.nature.com/articles/s41...
www.nature.com/articles/s41...
github.com/RosettaCommo...
You can now design proteins, and in particular enzymes from just partially defined amino acid side chains, and without defining their sequence position or order!

github.com/RosettaCommo...
You can now design proteins, and in particular enzymes from just partially defined amino acid side chains, and without defining their sequence position or order!

Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)

Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
SABER maps SaCas9 deletion tolerance for DNA binding, informing design of minimized CRISPRi repressors, revealing insights into SaCas9 structure.




SABER maps SaCas9 deletion tolerance for DNA binding, informing design of minimized CRISPRi repressors, revealing insights into SaCas9 structure.

www.nature.com/articles/s41...
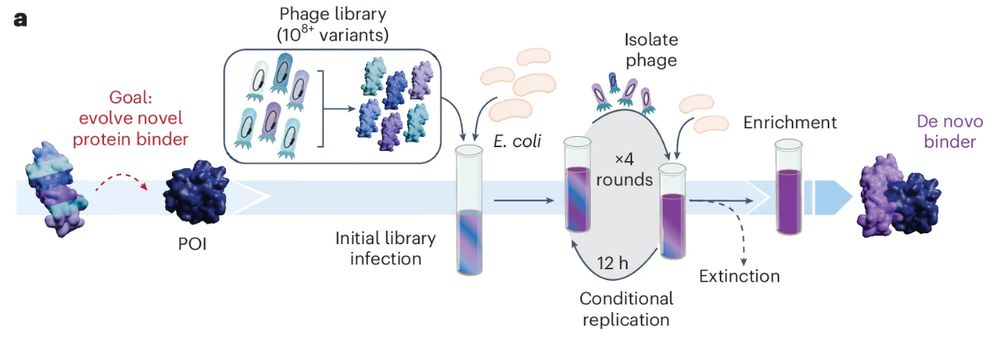
www.nature.com/articles/s41...

