

www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
A thread (1/11):
Thrilled to share our new work now published with Kathy Collins, @nogaleslab.bsky.social @berkeleymcb.bsky.social where we investigate this with #cryoEM & biochemistry in 🧪 and cells! #RNAsky #TEsky
Thrilled to share our new work now published with Kathy Collins, @nogaleslab.bsky.social @berkeleymcb.bsky.social where we investigate this with #cryoEM & biochemistry in 🧪 and cells! #RNAsky #TEsky
"I'm looking forward to the day that we are so slammed with work trying to reinstate everything that we had to terminate illegally — I'll work 24/7 to make that happen if I can."
"The explanations are bereft of reasoning — virtually in their entirety... unsupported by [facts]."
Each of them are VOID and ILLEGAL, he says.
"I'm looking forward to the day that we are so slammed with work trying to reinstate everything that we had to terminate illegally — I'll work 24/7 to make that happen if I can."


elifesciences.org/articles/77616

elifesciences.org/articles/77616




What a crazy innovative idea: Adapting the R2 retrotransposon to efficiently insert any custom transgene directly into RIBOSOMAL DNA ARRAYS! 🤯🤯🤯
The applications are endless....
rdcu.be/d8LVW

What a crazy innovative idea: Adapting the R2 retrotransposon to efficiently insert any custom transgene directly into RIBOSOMAL DNA ARRAYS! 🤯🤯🤯
The applications are endless....
onlinelibrary.wiley.com/doi/full/10....

onlinelibrary.wiley.com/doi/full/10....
As populations accumulated mutations, "some evolutionary paths that were inaccessible to the ancestor became accessible to the evolving populations, while others were closed off."
🦋🦫🌱🐋🧪 #philsci #evobio

www.nature.com/articles/s41...
20% of people assessed had accelerated aging in 1 organ, 1.7% multi-organ

www.nature.com/articles/s41...
20% of people assessed had accelerated aging in 1 organ, 1.7% multi-organ

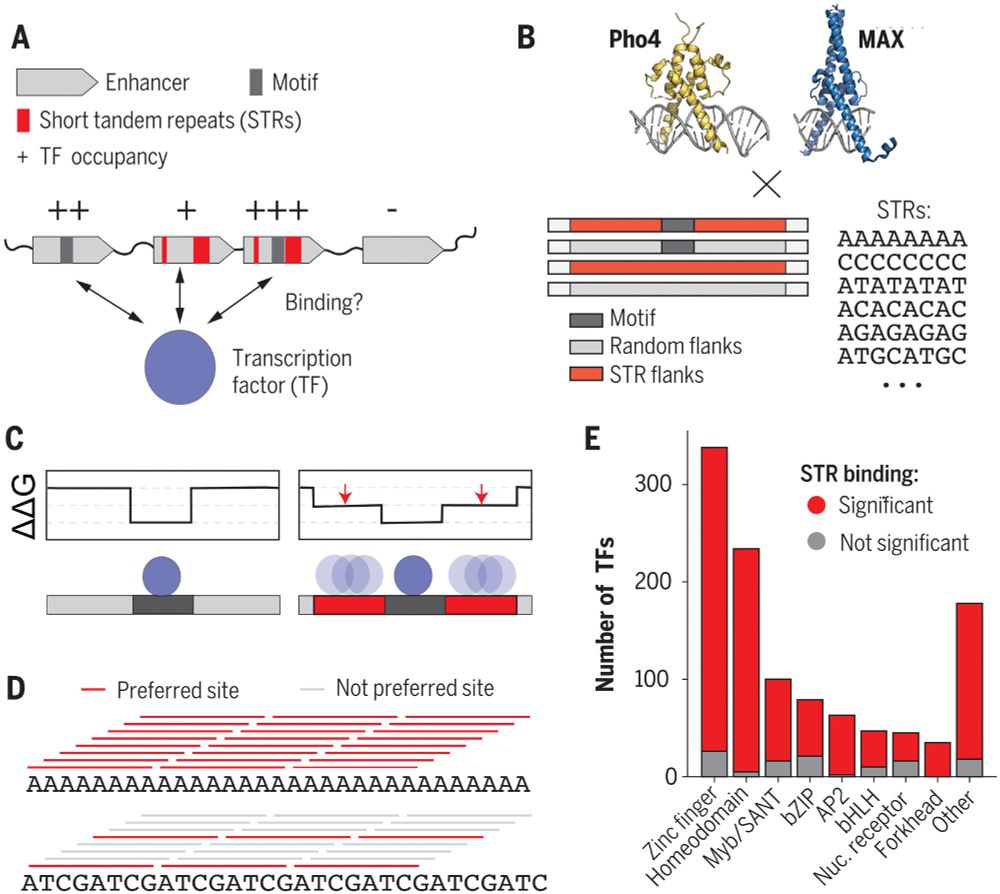
We do it all the time: fraserlab.com/reviews
The majority of the time, a journal asks us to include the review in their process AFTER we post!

We do it all the time: fraserlab.com/reviews
The majority of the time, a journal asks us to include the review in their process AFTER we post!
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
A thread (1/11):

www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
A thread (1/11):

