
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...
EMBL-EBI’s new AMR portal brings together laboratory resistance data and bacterial genomes in one open platform.
#WAAW2025 #ActOnAMR
www.ebi.ac.uk/about/news/t...
🧬💻

EMBL-EBI’s new AMR portal brings together laboratory resistance data and bacterial genomes in one open platform.
#WAAW2025 #ActOnAMR
www.ebi.ac.uk/about/news/t...
🧬💻
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.k.u-tokyo.ac.jp/en/informati...

www.k.u-tokyo.ac.jp/en/informati...
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
arxiv.org/abs/2506.12986

arxiv.org/abs/2506.12986
Unicore rapidly identifies structural single-copy core genes from input species proteomes for phylogenetic analysis. Powered by Foldseek and ProstT5, Unicore enables linear-scale structure-based phylogeny of any given set of taxa. 🧵1/n
📃 doi.org/10.1093/gbe/evaf109
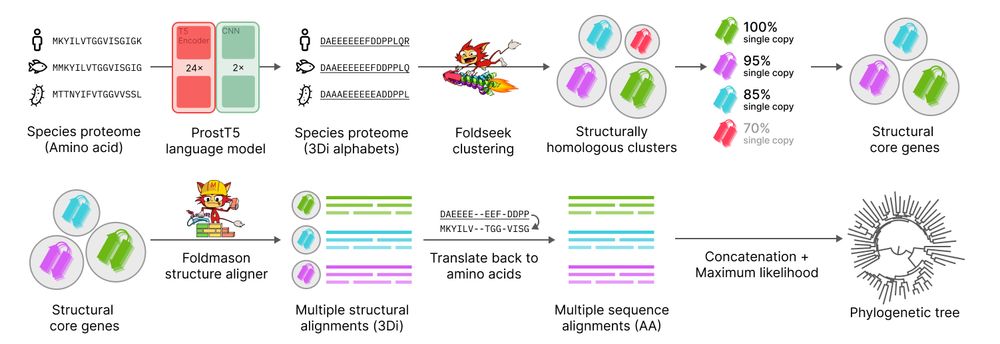
Unicore rapidly identifies structural single-copy core genes from input species proteomes for phylogenetic analysis. Powered by Foldseek and ProstT5, Unicore enables linear-scale structure-based phylogeny of any given set of taxa. 🧵1/n
📃 doi.org/10.1093/gbe/evaf109
The registrations for on-site & remote participation are still open! More info: RdRp.io
#RdRpSummit2025

The registrations for on-site & remote participation are still open! More info: RdRp.io
#RdRpSummit2025
Structural: MSAs, Virus DB, Core Genes, Motif Discovery, Multimer Clustering & Search, pLM Foldseek, Environmental analysis
Metagenomics: Classification & Metabuli App
GPU-based & RNA search, Proteome clustering, Novel Ribozyme discovery
& get Marv stickers!
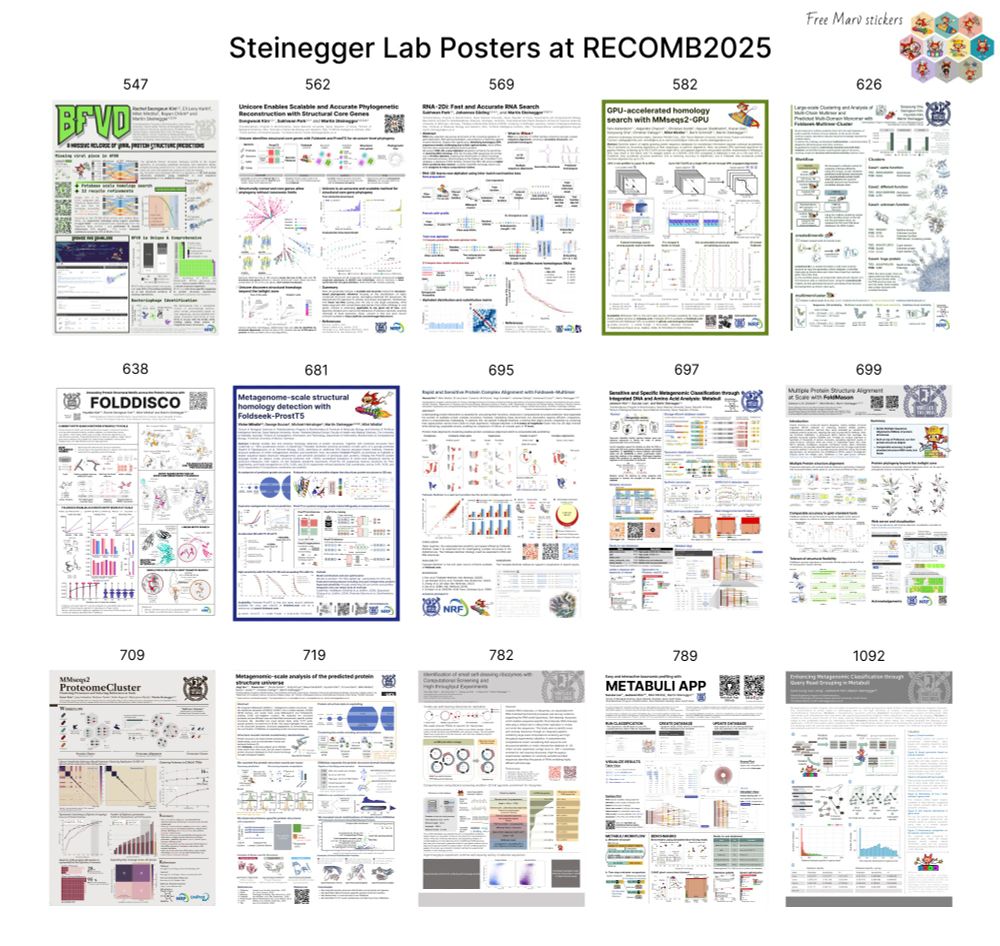
Structural: MSAs, Virus DB, Core Genes, Motif Discovery, Multimer Clustering & Search, pLM Foldseek, Environmental analysis
Metagenomics: Classification & Metabuli App
GPU-based & RNA search, Proteome clustering, Novel Ribozyme discovery
& get Marv stickers!

Structural: MSAs, Virus DB, Core Genes, Motif Discovery, Multimer Clustering & Search, pLM Foldseek, Environmental analysis
Metagenomics: Classification & Metabuli App
GPU-based & RNA search, Proteome clustering, Novel Ribozyme discovery
& get Marv stickers!
📄 academic.oup.com/nar/article/...
🌐 bfvd.foldseek.com
#RECOMB2025

📄 academic.oup.com/nar/article/...
🌐 bfvd.foldseek.com
#RECOMB2025
www.google.com/maps/d/edit?...

www.google.com/maps/d/edit?...
recomb.org/recomb2025/t...

recomb.org/recomb2025/t...

🌐https://bfvd.foldseek.com
💾https://bfvd.steineggerlab.workers.dev/
1/3
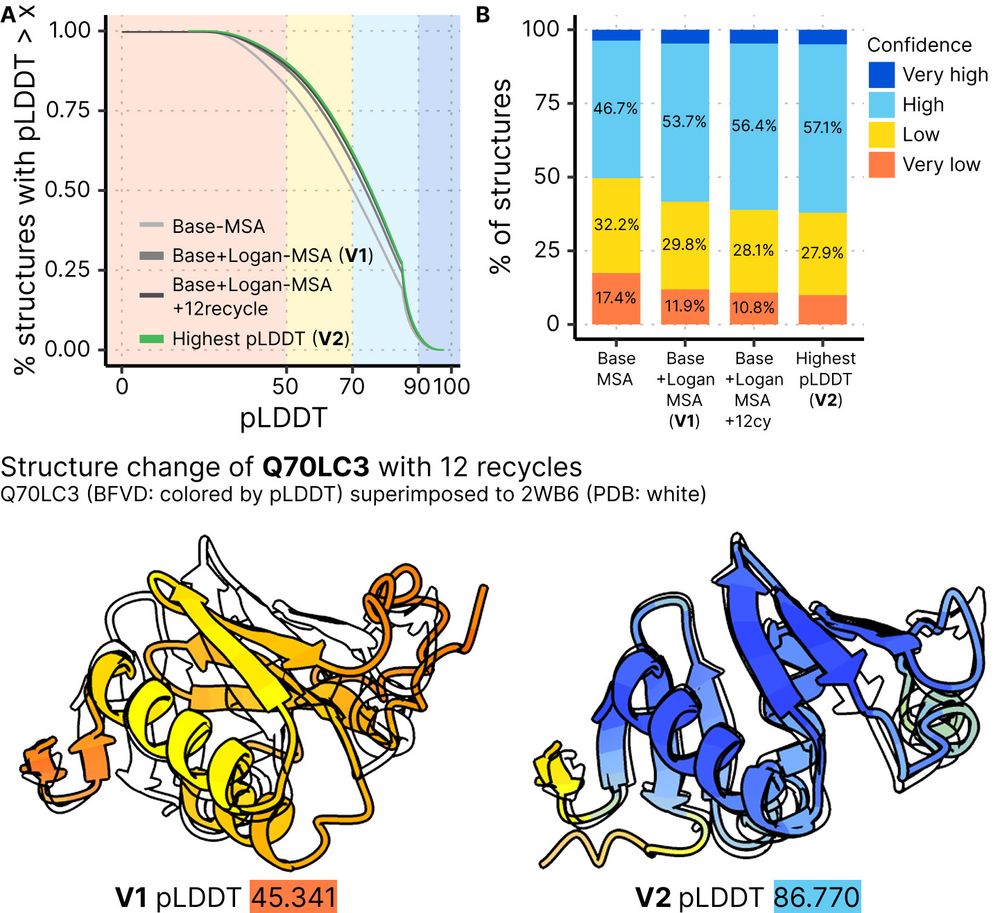
🌐https://bfvd.foldseek.com
💾https://bfvd.steineggerlab.workers.dev/
1/3
It includes quality-filtered GTDB R220, RefSeq viruses, and the human T2T.
Prokaryotes follow the GTDB taxonomy; viruses and the human genome use NCBI taxonomy.
Download it and expand with genomes of your choice using "updateDB" command.
metabuli.steineggerlab.workers.dev
It includes quality-filtered GTDB R220, RefSeq viruses, and the human T2T.
Prokaryotes follow the GTDB taxonomy; viruses and the human genome use NCBI taxonomy.
Download it and expand with genomes of your choice using "updateDB" command.
metabuli.steineggerlab.workers.dev
It's 7x to 30x faster than BinDash, by using the simd-minimizers crate for fast hashing, and a nearly branch-free implementation.
Here's a blogpost with a survey of minhash history & methods, and evals:
curiouscoding.nl/posts/simd-s...
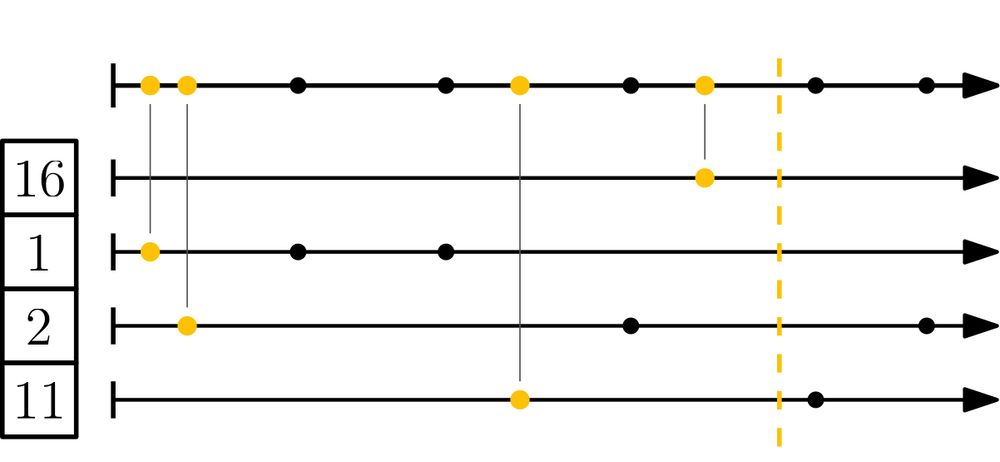
It's 7x to 30x faster than BinDash, by using the simd-minimizers crate for fast hashing, and a nearly branch-free implementation.
Here's a blogpost with a survey of minhash history & methods, and evals:
curiouscoding.nl/posts/simd-s...
💻Taxonomic classification & interactive visualization—right on your laptop
🛠️Create new databases or update existing ones with new sequences. 🧵1/5
github.com/steineggerla...
www.biorxiv.org/content/10.1101/2025.03.10.642298v1
💻Taxonomic classification & interactive visualization—right on your laptop
🛠️Create new databases or update existing ones with new sequences. 🧵1/5
github.com/steineggerla...
www.biorxiv.org/content/10.1101/2025.03.10.642298v1
📄 biorxiv.org/content/10.1...
💾 mmseqs.com and 🐍Bioconda 🖥️🧬🧶

📄 biorxiv.org/content/10.1...
💾 mmseqs.com and 🐍Bioconda 🖥️🧬🧶
🚀https://github.com/steineggerlab/Metabuli-App
🚀https://github.com/steineggerlab/Metabuli-App

