Base-pair resolution Micro Capture-C ultra to map chromatin contacts between individual motifs within cis-regulatory elements and reveal a unified model of biophysically mediated enhancer-promoter communication @cellcellpress.bsky.social
www.cell.com/cell/fulltex...
www.cell.com/cell/fulltex...
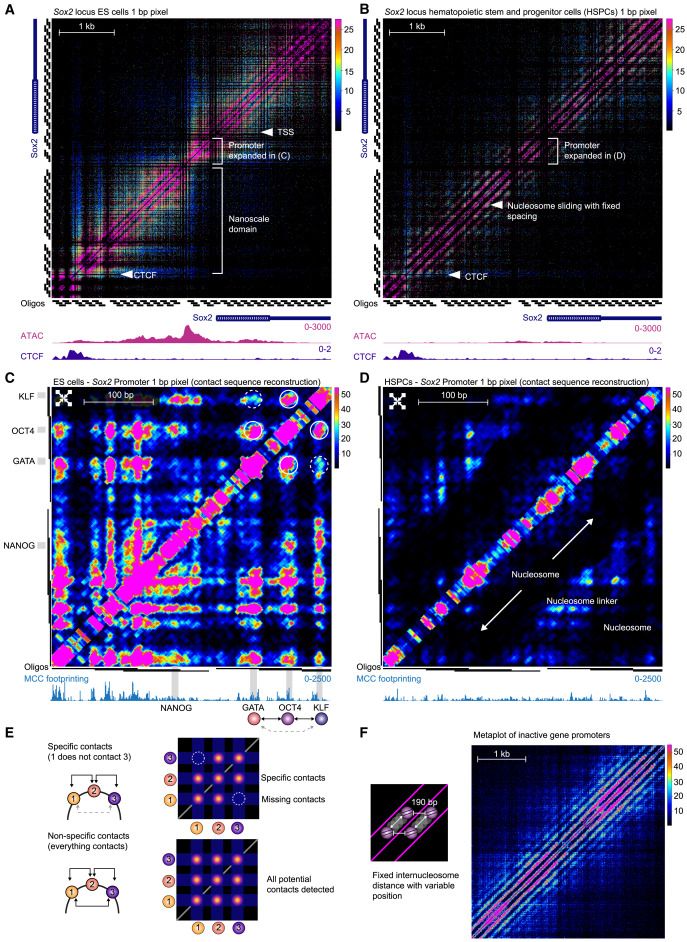
November 6, 2025 at 3:03 AM
Everybody can reply
2 reposts
7 likes
2 saves
50 years ago, King & Wilson published a foundational paper that underlies the cis-regulatory paradigm (CRP) of #DevoEvo #EvoDevo, i.e., that *almost* all morphological evolution is driven by mutations in regulatory elements, rather than proteins, and it all arose from simple misunderstanding 🧪 🧵
October 29, 2025 at 12:35 AM
Everybody can reply
32 reposts
5 quotes
70 likes
21 saves
🚀 New preprint!
We introduce K-PROB, a k-mer–based Bayesian framework that mines pangenomic promoter and expression diversity to predict cis-regulatory elements driving gene expression variation in crops 🌽🌱
🔗 biorxiv.org/content/10.1101/2025.10.27.684862v1
#Genomics #GeneRegulation #Maize #Soybean
We introduce K-PROB, a k-mer–based Bayesian framework that mines pangenomic promoter and expression diversity to predict cis-regulatory elements driving gene expression variation in crops 🌽🌱
🔗 biorxiv.org/content/10.1101/2025.10.27.684862v1
#Genomics #GeneRegulation #Maize #Soybean

Natural variation in regulatory code revealed through Bayesian analysis of plant pan-genomes and pan-transcriptomes
Understanding the genetic code of cis-regulatory elements (CREs) is essential for engineering gene expression and modulating agronomic traits in crops. In plants, CREs underlying rapid evolution of ge...
biorxiv.org
October 29, 2025 at 4:47 PM
Everybody can reply
17 reposts
32 likes
3 saves
It was such a pleasure to host @joadelas.bsky.social at @edinuni-irr.bsky.social yesterday. What a great talk- fascinating insights into how Cis Regulatory Elements process spatial and temporal information to build complicated and beautiful things such as embryos. Bravo Joaquina!
Go to sleep in London, wake up in Edinburg! Great experience aboard the Caledonian Sleeper train.
Looking forward to a fun day full of science at the CRM. Thank you so much @cellysally.bsky.social for hosting me.
Looking forward to a fun day full of science at the CRM. Thank you so much @cellysally.bsky.social for hosting me.
October 31, 2025 at 9:06 AM
Everybody can reply
6 likes
The original sin of the CRP is its argument that changes in regulatory proteins (transcription factors) would have deleterious pleiotropic effects while changes in modular cis-regulatory elements would not, thus morphological evolution almost always results of mutations in regulatory elements...
October 29, 2025 at 12:35 AM
Everybody can reply
4 likes
Non-recurrent duplications on chromosome 4p16.1 involving cis-regulatory elements affecting neural crest development in patients with isolated bilateral microtia #RareDisease #Genetics link.springer.com/article/10.1...

Non-recurrent duplications on chromosome 4p16.1 involving cis-regulatory elements affecting neural crest development in patients with isolated bilateral microtia - Human Genetics
Microtia-anotia is a common congenital anomaly. In most cases, the genetic etiology remains unknown. The proper development of outer ear is closely related to cranial neural crest cells. Abnormal DNA ...
link.springer.com
October 29, 2025 at 2:58 PM
Everybody can reply
1 likes
It is, but rationale one at the time, it's philosophical and so unanswerable: what is the definition of importance? The King and Wilson paper is always cited as the theoretical foundation of the cis-regulatory paradigm; my real point is that that’s wrong; Wilson (at least) was much more broad...
October 29, 2025 at 2:46 PM
Everybody can reply
1 likes
Guo et al. generated haplotype-resolved genome assemblies of black carp, identifying dmrt1 as the sex-determining locus, with a 13.4 kb insertion containing TEs functioning as cis-regulatory modules that mediate Y-specific activation.
🔗 doi.org/10.1093/molbev/msaf239
#evobio #molbio #TEsky
🔗 doi.org/10.1093/molbev/msaf239
#evobio #molbio #TEsky

Transposable Element-Mediated Cis-Regulation Drives the Evolution of dmrt1 as a Candidate Master Sex-Determining Gene in Black Carp
Abstract. Sex determination in vertebrates exhibits remarkable evolutionary plasticity, with diverse mechanisms and master sex-determining (MSD) genes aris
doi.org
October 21, 2025 at 9:47 AM
Everybody can reply
4 reposts
11 likes
3 saves
even more results in the #preprint!
Our interpretation (TL;DR)
We think that the promoter-proximal cis-regulatory element acts like a #facilitator that helps to filter and relay long-range enhancer activity to the core promoter!
Our interpretation (TL;DR)
We think that the promoter-proximal cis-regulatory element acts like a #facilitator that helps to filter and relay long-range enhancer activity to the core promoter!
October 16, 2025 at 3:06 PM
Everybody can reply
2 reposts
7 likes
What is a promoter and how does it work? Let’s dive in!
We wanted to understand why most promoters have some sort of cis-regulatory element (a.k.a. promoter-proximal region a.k.a. proximal enhancer) directly upstream of the core promoter.
We wanted to understand why most promoters have some sort of cis-regulatory element (a.k.a. promoter-proximal region a.k.a. proximal enhancer) directly upstream of the core promoter.

October 16, 2025 at 3:06 PM
Everybody can reply
3 reposts
6 likes
More questions! If Sox9 has so many long-range enhancers - Can core promoters alone be activated without #promoter-proximal-region-proximalenhancer-cis-regulatory-element?
So we tested only core promoters in our assay
FACT3: No - core promoters alone are not activated by Sox9 long-range enhancers
So we tested only core promoters in our assay
FACT3: No - core promoters alone are not activated by Sox9 long-range enhancers

October 16, 2025 at 3:06 PM
Everybody can reply
3 likes
New Online! Cis-regulatory elements that tune transcriptional responses in liver drug metabolism and outcomes

Cis-regulatory elements that tune transcriptional responses in liver drug metabolism and outcomes
Nature Reviews Molecular Cell Biology, Published online: 10 October 2025; doi:10.1038/s41580-025-00916-2The discovery of a cis-regulatory element required for xenobiotic gene activation highlighted the crucial role of enhancers in drug metabolism.
bit.ly
October 10, 2025 at 2:35 PM
Everybody can reply
1 likes
Which mutations rewire function of regulatory DNA?
Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇
Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇

October 9, 2025 at 12:03 PM
Everybody can reply
10 reposts
28 likes
2 saves
I’m excited to share our new preprint! We built a single-nucleus multiomic (snRNA-seq+snATAC-seq) atlas of leptin receptor-expressing hypothalamic neurons (LepRᴴʸᵖᵒ) and identified conserved cis-regulatory elements overlapping human obesity-linked variants and eQTLs🧠🧬
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Single-nucleus transcriptional and chromatin accessibility profiling of mouse hypothalamic LepRb neurons reveals cell type-specific cis-regulatory elements linked to human obesity
Leptin receptor-expressing hypothalamic neurons (LepRHypo) are key regulators of energy balance, yet a comprehensive, cell type-resolved, chromatin accessibility map of these neurons is lacking. We pr...
www.biorxiv.org
October 8, 2025 at 5:02 PM
Everybody can reply
7 reposts
26 likes
1 saves
ICYMI: New online: Reporter CRISPR screens decipher cis-regulatory and trans-regulatory principles at the Xist locus

Reporter CRISPR screens decipher cis-regulatory and trans-regulatory principles at the Xist locus
Nature Structural & Molecular Biology, Published online: 06 October 2025; doi:10.1038/s41594-025-01686-3Here Schwämmle et al. develop CRISPR reporter screens to map transcription-factor-regulatory element interactions at the Xist locus, revealing a two-step mechanism integrating developmental and X-dosage signals to initiate X-chromosome inactivation.
go.nature.com
October 7, 2025 at 12:36 PM
Everybody can reply
1 likes
Happy to share work spearheaded by former grad student Colin Shew testing shared duplicated cis regulatory elements (CREs) using an MPRA. While we find some high effect CREs, collectively paralog differences represent modest effects accounting for observed gene expression divergence.
Influence of cis-regulatory elements on regulatory divergence in human segmental duplications https://www.biorxiv.org/content/10.1101/2025.10.03.680410v1
October 7, 2025 at 2:31 AM
Everybody can reply
13 reposts
30 likes
Inferring gene-regulatory networks using epigenomic priors [updated]
Epigenomic priors enhance GRN inference by leveraging DNA methylation data. This identifies differential cis-regulation patterns relevant to development/cancer.
Epigenomic priors enhance GRN inference by leveraging DNA methylation data. This identifies differential cis-regulation patterns relevant to development/cancer.




October 7, 2025 at 10:41 AM
Everybody can reply
1 likes
Influence of cis-regulatory elements on regulatory divergence in human segmental duplications https://www.biorxiv.org/content/10.1101/2025.10.03.680410v1
October 5, 2025 at 5:33 PM
Everybody can reply
1 quotes
2 likes
3 saves
ICYMI: New online! Cis-regulatory elements at cellular resolution
Cis-regulatory elements at cellular resolution
Nature Reviews Genetics, Published online: 16 September 2025; doi:10.1038/s41576-025-00882-7In this Comment, the authors outline some key next steps to advance our understanding of cis-regulatory elements at single-cell resolution, which includes harmonizing global efforts to construct a comprehensive single-cell atlas of gene regulation.
www.nature.com
September 23, 2025 at 3:32 PM
Everybody can reply
4 likes

A deep-time landscape of plant cis-regulatory sequence evolution
Developmental gene function is conserved over deep time, but cis-regulatory sequence conservation is rarely found. Rapid sequence turnover, paleopolyploidy, structural variation, and limited phylogeno...
www.biorxiv.org
September 22, 2025 at 12:54 AM
Everybody can reply
4 reposts
18 likes
3 saves
“These results highlight how adaptive phenotypes may be more mutationally accessible in coding sequences than promoters, especially when large effects are beneficial.”
Ha, take that Cis-Regulatory Paradigm! #DevoEvo #EvoDevo 🧪
Ha, take that Cis-Regulatory Paradigm! #DevoEvo #EvoDevo 🧪
Really excited to share our latest paper led by @simonaube.bsky.social. Fascinating results examining whether regulatory mutations can lead to adaptation as fast as coding mutations do. #mevosky #evobio #evoSky
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Adaptation is less accessible through mutations in promoters than in coding sequences when large effect sizes are needed
Mutations within promoters and coding sequences are both involved in adaptation, but their relative contributions remain to be compared directly. Using the fungal enzyme cytosine deaminase, we examine...
www.biorxiv.org
September 12, 2025 at 11:16 PM
Everybody can reply
2 reposts
16 likes
Petty excited about this work with Alan Moses on power of predicting cis regulatory gene networks www.biorxiv.org/content/10.1...

Inferring fungal cis-regulatory networks from genome sequences
Gene expression patterns are determined to a large extent by transcription factor binding to non-coding regulatory regions in the genome. However, gene expression cannot yet be systematically predicte...
www.biorxiv.org
March 4, 2025 at 9:41 AM
Everybody can reply
4 reposts
12 likes
Identifying non-coding variant effects at scale via machine learning models of cis-regulatory reporter assays https://www.biorxiv.org/content/10.1101/2025.04.16.648420v1
April 18, 2025 at 4:32 PM
Everybody can reply
very excited to share the last chunk of my PhD work, now available on @biorxivpreprint.bsky.social! in this work, we explore how cis-regulatory variation and gene-environment (GxE) interactions influence immune response diversity directly in a cohort of hospitalized COVID-19 patients 1/n
Widespread gene-environment interactions shape the immune response to SARS-CoV-2 infection in hospitalized COVID-19 patients https://www.biorxiv.org/content/10.1101/2024.12.03.626676v1

Widespread gene-environment interactions shape the immune response to SARS-CoV-2 infection in hospitalized COVID-19 patients https://www.biorxiv.org/content/10.1101/2024.12.03.626676v1
Genome-wide association studies performed in patients with coronavirus disease 2019 (COVID-19) have
www.biorxiv.org
December 6, 2024 at 9:46 PM
Everybody can reply
3 reposts
12 likes
Cis-regulatory polymorphism at fiz ecdysone oxidase contributes to polygenic adaptation to malnutrition in Drosophila https://www.biorxiv.org/content/10.1101/2023.08.28.555138v1

Cis-regulatory polymorphism at fiz ecdysone oxidase contributes to polygenic adaptation to malnutrition in Drosophila https://www.biorxiv.org/content/10.1101/2023.08.28.555138v1
We investigate the contribution of a candidate gene, fiz (fezzik), to complex polygenic adaptation t
www.biorxiv.org
August 30, 2023 at 12:33 AM
Everybody can reply

