Interested in ecology and evolution of microbial communities, microbiome, culturomics, experimental (co)evolution, microbiome modulation
🦠🧫🔬🧪💊💩🧬
https://scholar.google.com/citations?user=dhZfxqMAA
Check it out here:
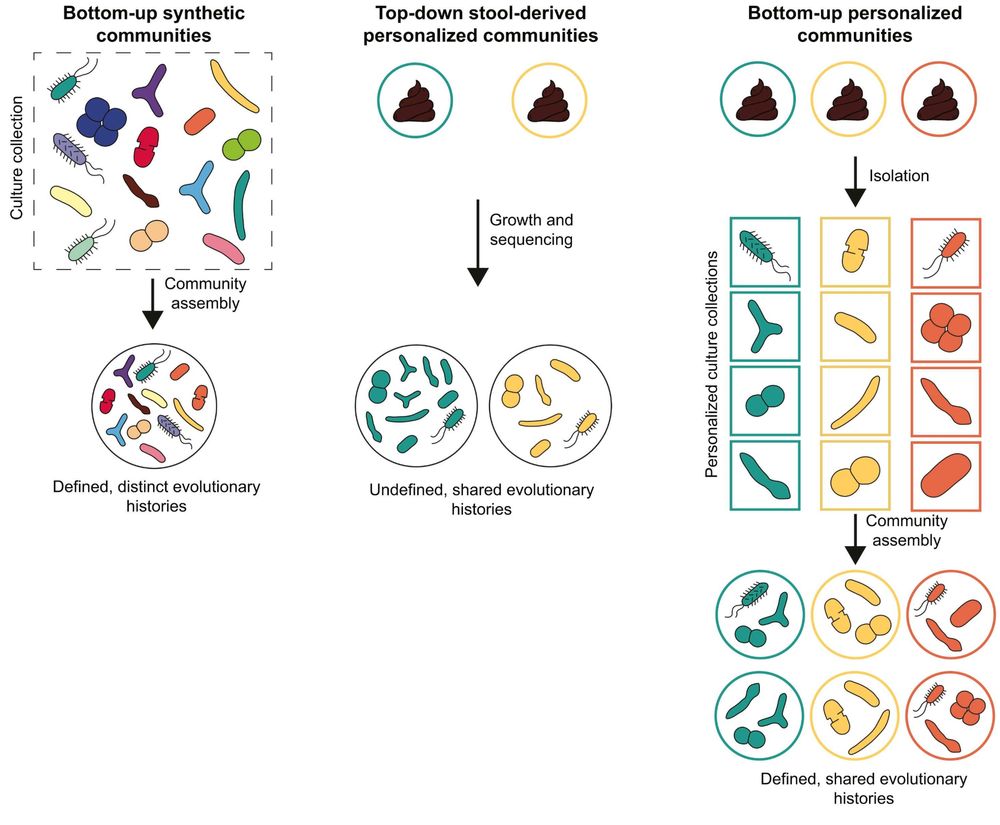
Harnessing gut microbial communities to unravel microbiome functions
doi.org/10.1016/j.mi...
@typaslab.bsky.social
www.science.org/doi/full/10....

www.science.org/doi/full/10....

This work has so many implications on literature 😳😬
Fluorescein-based dyes are not valid reporters of oxidative stress in bacteria, and conclusions based on their use must be reconsidered | PNAS www.pnas.org/doi/10.1073/...

This work has so many implications on literature 😳😬
Fluorescein-based dyes are not valid reporters of oxidative stress in bacteria, and conclusions based on their use must be reconsidered | PNAS www.pnas.org/doi/10.1073/...


www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...

www.pnas.org/doi/10.1073/...
Sadly, the Editors at PNAS rejected our initial introduction, which was a David Attenborough style voice over of the microbial Serengeti (included below)

www.pnas.org/doi/10.1073/...
Sadly, the Editors at PNAS rejected our initial introduction, which was a David Attenborough style voice over of the microbial Serengeti (included below)
pmc.ncbi.nlm.nih.gov/articles/PMC...
You can read our news and views here: www.pnas.org/doi/10.1073/...
pmc.ncbi.nlm.nih.gov/articles/PMC...
You can read our news and views here: www.pnas.org/doi/10.1073/...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Defensive fungal symbiosis on insect hindlegs: www.science.org/doi/10.1126/...
#SymbioSky

Defensive fungal symbiosis on insect hindlegs: www.science.org/doi/10.1126/...
#SymbioSky
www.pnas.org/doi/10.1073/...

www.pnas.org/doi/10.1073/...
Supportive and frequent supervision is a key factor that significantly enhances PhD students' satisfaction and well-being during their research journey.
doi.org/10.1038/d415...

Supportive and frequent supervision is a key factor that significantly enhances PhD students' satisfaction and well-being during their research journey.
doi.org/10.1038/d415...
www.nature.com/articles/s41...
🧵⬇️
1/8

www.nature.com/articles/s41...
🧵⬇️
1/8

@natmicrobiol.nature.com today!
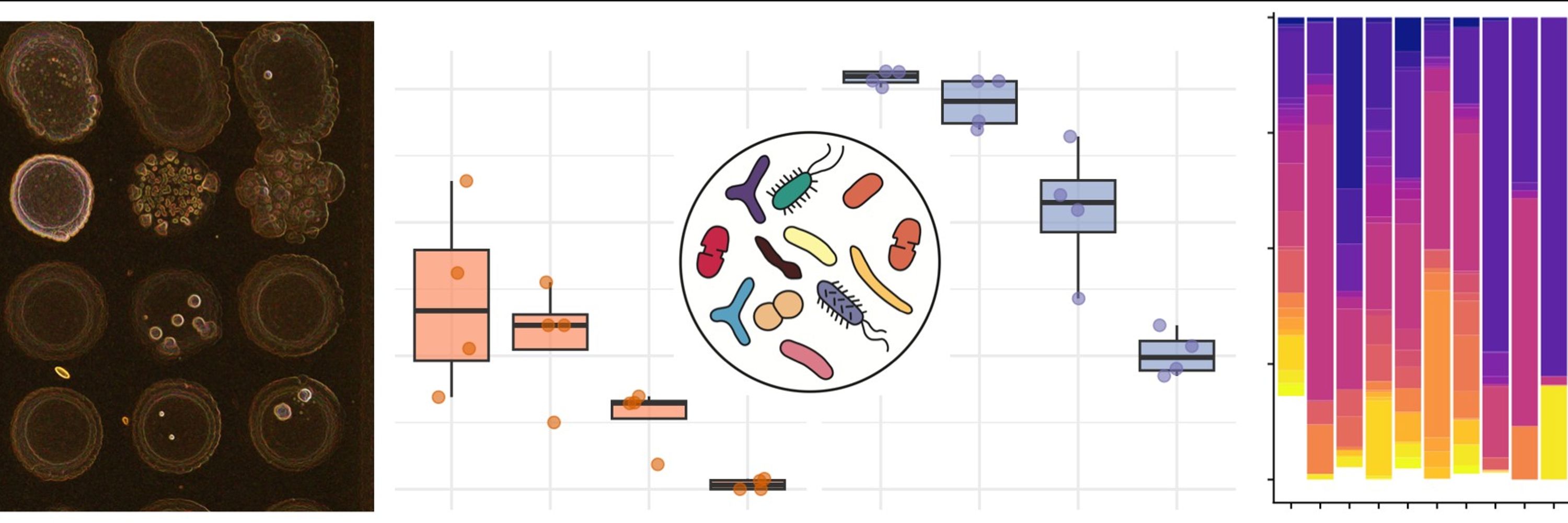
Fecal microbiota transplant (FMT), donor 💩 => patients' gut, is an effective treatment for recurrent C. difficile infection & is being evaluated for Inflammatory Bowel Diseases (IBD) & other conditions 1/
📄 rdcu.be/eL8mR

@natmicrobiol.nature.com today!
Fecal microbiota transplant (FMT), donor 💩 => patients' gut, is an effective treatment for recurrent C. difficile infection & is being evaluated for Inflammatory Bowel Diseases (IBD) & other conditions 1/
📄 rdcu.be/eL8mR
academic.oup.com/nar/advance-...

academic.oup.com/nar/advance-...
rdcu.be/eMcNg

rdcu.be/eMcNg
Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...

Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...
jobs.awi.de/Vacancies/20...
This is quite an exciting opportunity to push the boundaries of what is known regarding the molecular basis of the formation and demise of photosymbiotic relationships in marine habitats.
jobs.awi.de/Vacancies/20...
This is quite an exciting opportunity to push the boundaries of what is known regarding the molecular basis of the formation and demise of photosymbiotic relationships in marine habitats.
nature.com/articles/s41...
PhD graduate and now post-doc Sofia Dahlman, along with co-senior author Sam Forster from The Hudson and other researchers from our lab and others.

nature.com/articles/s41...
PhD graduate and now post-doc Sofia Dahlman, along with co-senior author Sam Forster from The Hudson and other researchers from our lab and others.



