- phages, microbes and more

www.biorxiv.org/content/10.1...




Thanks everyone who joined the workshop on Orchestrating Microbiome Analysis with Bioconductor, you made it awesome!
@abacbs.bsky.social
@bioconductor.bsky.social
#ABACBS2025
#BioCAsia2025

Thanks everyone who joined the workshop on Orchestrating Microbiome Analysis with Bioconductor, you made it awesome!
@abacbs.bsky.social
@bioconductor.bsky.social
#ABACBS2025
#BioCAsia2025
I came out of it with a Wasm app for making pharokka/phold/phynteny's genome maps - available at gbouras13.github.io/phold-plot-w...
It runs the browser almost instantly - please give it a go with your phages of interest!
#ABACBS2025
I came out of it with a Wasm app for making pharokka/phold/phynteny's genome maps - available at gbouras13.github.io/phold-plot-w...
It runs the browser almost instantly - please give it a go with your phages of interest!
#ABACBS2025

#bioinformatics #graphs #graph-algorithms #flow-decomposition #integer-linear-programming
github.com/algbio/flowp...
#bioinformatics #graphs #graph-algorithms #flow-decomposition #integer-linear-programming
github.com/algbio/flowp...




It can do near-T2T assembly using @nanoporetech.com adaptive sampling
- with less 💸
- reference agnostic, so works for non-humans
- not just blood, even saliva
Just presented at #abacbs2025 yesterday.
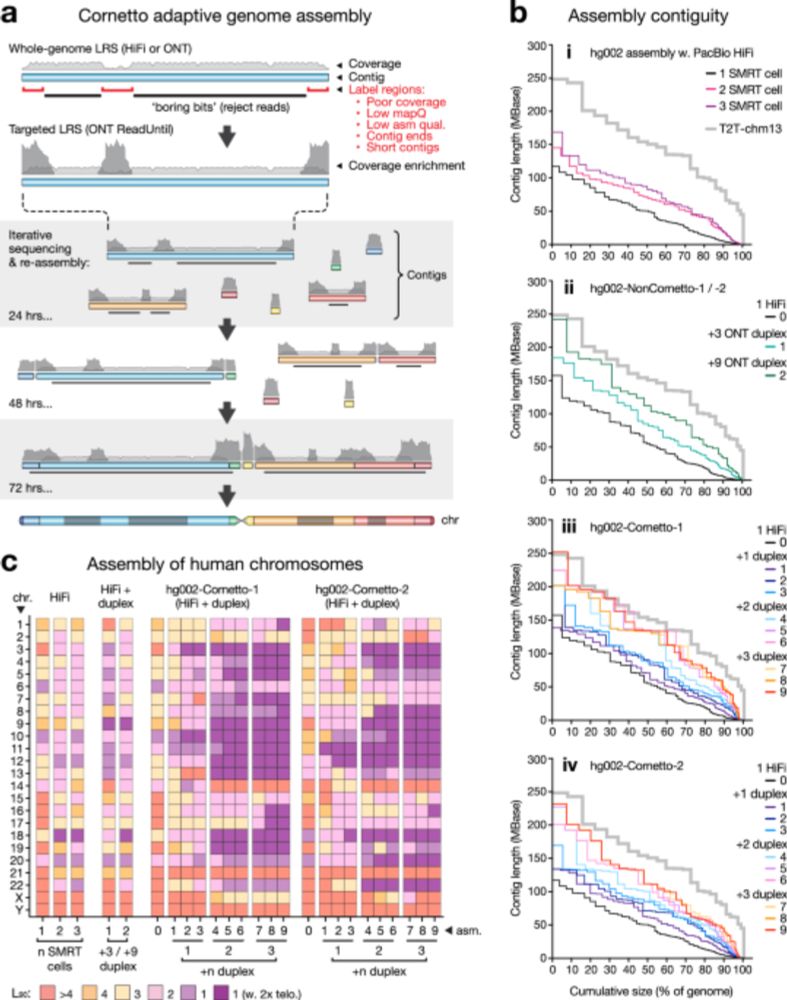
It can do near-T2T assembly using @nanoporetech.com adaptive sampling
- with less 💸
- reference agnostic, so works for non-humans
- not just blood, even saliva
Just presented at #abacbs2025 yesterday.




#phagesky
www.nature.com/articles/s41...








#ABACBS2025
⚫️🔴
🔵⚪️

#ABACBS2025
⚫️🔴
🔵⚪️

#Confbingo update - presentation on dangerous animals ✅✅ #ABACBS2025



#Confbingo update - presentation on dangerous animals ✅✅ #ABACBS2025

Precision and Variant Calling in Acute Lymphoblastic Leukaemia
Patients

Precision and Variant Calling in Acute Lymphoblastic Leukaemia
Patients

#ABACBS2025
⚫️🔴
🔵⚪️

#ABACBS2025
⚫️🔴
🔵⚪️
PhD candidate @bonson-wong.bsky.social (poster at #abacbs2025) and great to see being highlighted in the AMD blog: www.amd.com/en/blogs/202...
PhD candidate @bonson-wong.bsky.social (poster at #abacbs2025) and great to see being highlighted in the AMD blog: www.amd.com/en/blogs/202...
doi.org/10.1101/2025...

doi.org/10.1101/2025...



