#phagesky
www.nature.com/articles/s41...

#phagesky
www.nature.com/articles/s41...

#phagesky
www.nature.com/articles/s41...
•
💻 Github: github.com/Vini2/agtools
📄 Preprint: biorxiv.org/content/10.110…

•
💻 Github: github.com/Vini2/agtools
📄 Preprint: biorxiv.org/content/10.110…
It can do near-T2T assembly using @nanoporetech.com adaptive sampling
- with less 💸
- reference agnostic, so works for non-humans
- not just blood, even saliva
Just presented at #abacbs2025 yesterday.
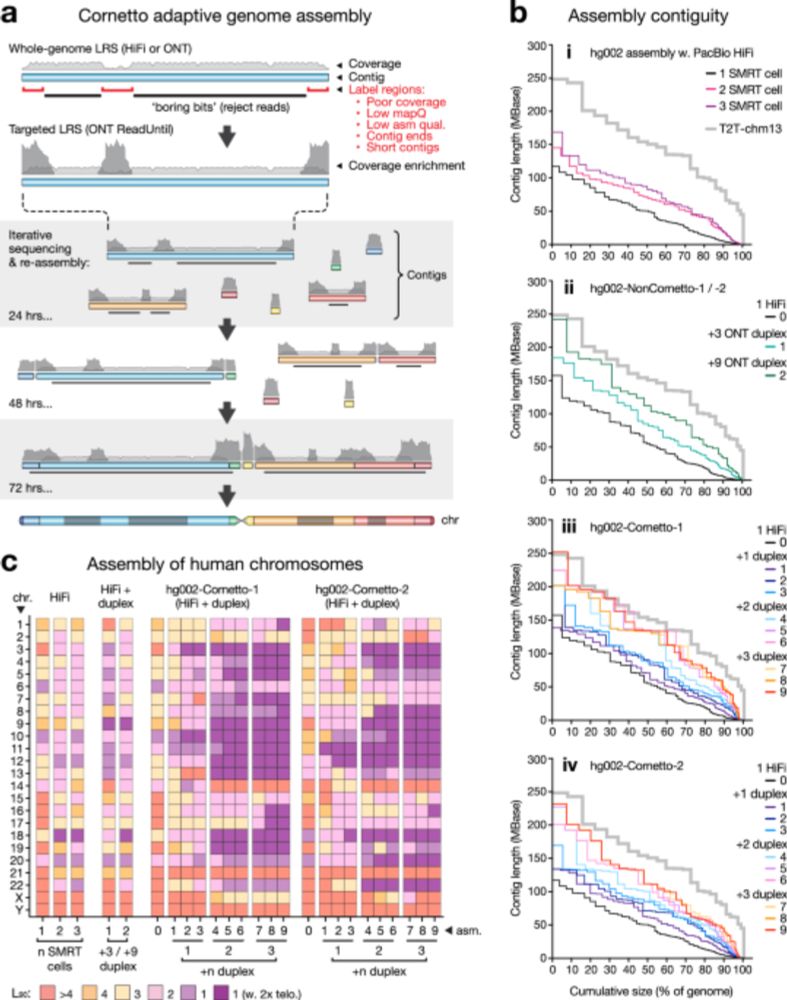
It can do near-T2T assembly using @nanoporetech.com adaptive sampling
- with less 💸
- reference agnostic, so works for non-humans
- not just blood, even saliva
Just presented at #abacbs2025 yesterday.



#ABACBS2025
⚫️🔴
🔵⚪️

#ABACBS2025
⚫️🔴
🔵⚪️

Come find my poster #21.
#ABACBS2025 @abacbs.bsky.social

Come find my poster #21.
#ABACBS2025 @abacbs.bsky.social
PhD candidate @bonson-wong.bsky.social (poster at #abacbs2025) and great to see being highlighted in the AMD blog: www.amd.com/en/blogs/202...
PhD candidate @bonson-wong.bsky.social (poster at #abacbs2025) and great to see being highlighted in the AMD blog: www.amd.com/en/blogs/202...











@aliciao.bsky.social @nadia-davidson.bsky.social @shazanfar.bsky.social @ellispatrick.bsky.social Jean Yang Belinda Phipson Milica Ng

@aliciao.bsky.social @nadia-davidson.bsky.social @shazanfar.bsky.social @ellispatrick.bsky.social Jean Yang Belinda Phipson Milica Ng

Sequence data production is exponential!
Dr Joanna McEntyre, EMBL-EBI
#ABACBS2025
Sequence data production is exponential!
Dr Joanna McEntyre, EMBL-EBI
#ABACBS2025
Exabytes of data but the biggest growth nowadays is imaging data
Dr Joanna McEntyre. Interim Director, EMBL-EBI
#ABACBS2025

Exabytes of data but the biggest growth nowadays is imaging data
Dr Joanna McEntyre. Interim Director, EMBL-EBI
#ABACBS2025
I am very excited to finally share what has been the main focus of my PhD for the past almost 3 years! It is about viral dark matter and a powerful tool we built to shed light on it. 🧬💡
Continue reading (🧵)

#phage #ABACBS2025

#phage #ABACBS2025

