
#TPD #ChemSky #ChemBio

#TPD #ChemSky #ChemBio
According to this new study by the @sebastianpomplun.bsky.social group of our LED3 hub, you may even get away without needing the DNA barcode. They describe self-encoded libraries (SELs) for #DrugDiscovery.
www.nature.com/articles/s41...
#ChemSky #ChemBio

According to this new study by the @sebastianpomplun.bsky.social group of our LED3 hub, you may even get away without needing the DNA barcode. They describe self-encoded libraries (SELs) for #DrugDiscovery.
www.nature.com/articles/s41...
#ChemSky #ChemBio
py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)

py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)

A big leap in understanding TB’s survival tricks and new angles for therapies.
📖 shorturl.at/Z8MVX
✍️ @genevauxpierre.bsky.social & coll.
#MicroSky

A big leap in understanding TB’s survival tricks and new angles for therapies.
📖 shorturl.at/Z8MVX
✍️ @genevauxpierre.bsky.social & coll.
#MicroSky
#ChemSky #ChemBio #DrugDiscovery #ABPP
www.nature.com/articles/s41...
#ChemSky #ChemBio #DrugDiscovery #ABPP
www.nature.com/articles/s41...
www.jnj.com/media-center...
www.jnj.com/media-center...
1. Introduced a broad definition of “biomolecular condensates,” promoting acceptance that a unifying mechanism may underlie all membrane-less compartments.

1. Introduced a broad definition of “biomolecular condensates,” promoting acceptance that a unifying mechanism may underlie all membrane-less compartments.
onlinelibrary.wiley.com/doi/10.1002/...

onlinelibrary.wiley.com/doi/10.1002/...


#chemsky 🧪

#chemsky 🧪

DeGrado, Arikin, Picotti (Keynotes). @lmkdassama.bsky.social @brianliau.bsky.social @rhodamine110.bsky.social @benlehner.bsky.social @alitavassoli.bsky.social
www.embl.org/about/info/c...
DeGrado, Arikin, Picotti (Keynotes). @lmkdassama.bsky.social @brianliau.bsky.social @rhodamine110.bsky.social @benlehner.bsky.social @alitavassoli.bsky.social
www.embl.org/about/info/c...
doi.org/10.1038/s414...

doi.org/10.1038/s414...
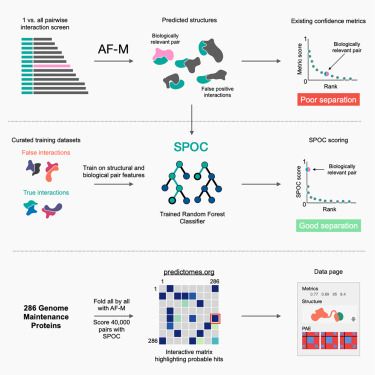
Cytosolic acetyl-coenzyme A is a signalling metabolite to control mitophagy

Cytosolic acetyl-coenzyme A is a signalling metabolite to control mitophagy
We obtained lots of thermal stable plastic degrading enzymes from the deep sea (Guaymas Basin, Gulf of California)

We obtained lots of thermal stable plastic degrading enzymes from the deep sea (Guaymas Basin, Gulf of California)
bit.ly/3VMcyNZ

bit.ly/3VMcyNZ

#ChemSky #ChemBio #TPD


#ChemSky #ChemBio #TPD



