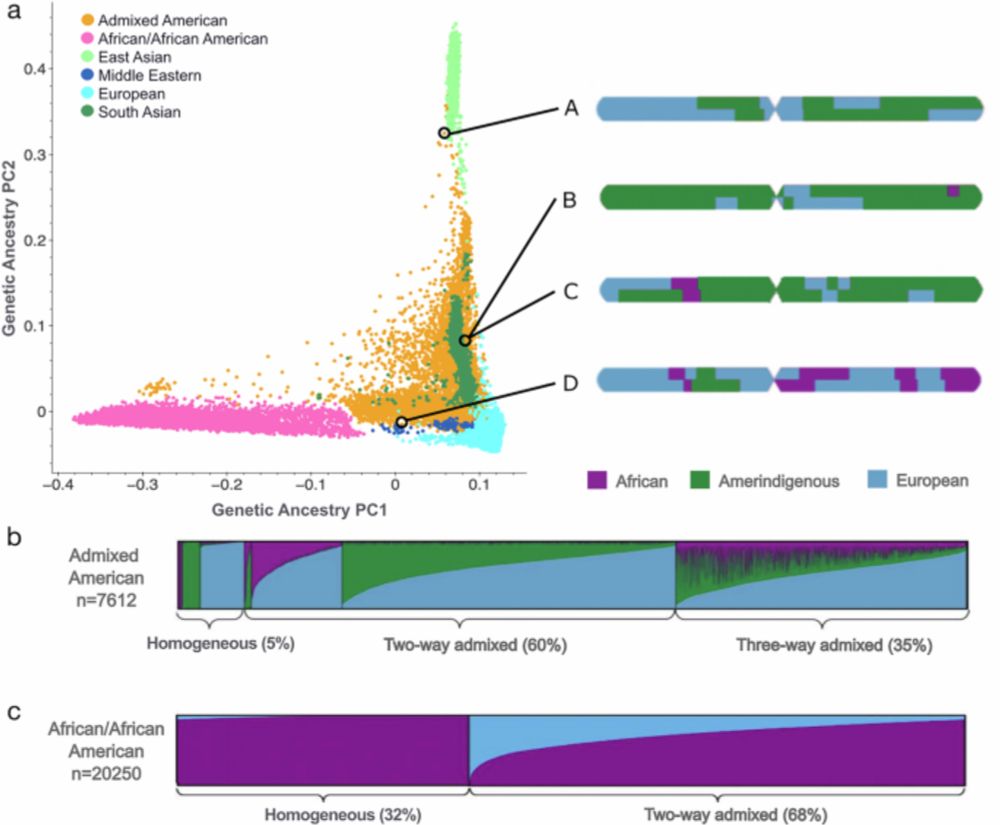
Science: disorder, condensates, repeats, cell stress, neurodegeneration, drug discovery, synbio Non-science: art, fashion, cooking
www.boeynaemslab.org
#protistsonsky 🧵
#protistsonsky 🧵
Study led by Ved Topkar brilliant MD/PhD student who used biochemistry and bioinformatics to probe the relationship between Mbp mRNA structure & transport/local translation in oligodendrocytes!
biorxiv.org/content/10.110…
1/8
Study led by Ved Topkar brilliant MD/PhD student who used biochemistry and bioinformatics to probe the relationship between Mbp mRNA structure & transport/local translation in oligodendrocytes!
biorxiv.org/content/10.110…
1/8


www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

Or from a single dish of HEK cell culture in the case of two membrane proteins.
Out in Nature Methods now! lnkd.in/gpyBSceg
Wonderful collaboration with the Efremov lab.

Or from a single dish of HEK cell culture in the case of two membrane proteins.
Out in Nature Methods now! lnkd.in/gpyBSceg
Wonderful collaboration with the Efremov lab.
Brilliant study led by @fmacleod.bsky.social and Andriko von Kügelgen. Tight collaboration with @buzzbaum.bsky.social and lab. Congrats to all authors!
www.biorxiv.org/content/10.1...

Brilliant study led by @fmacleod.bsky.social and Andriko von Kügelgen. Tight collaboration with @buzzbaum.bsky.social and lab. Congrats to all authors!
www.biorxiv.org/content/10.1...
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.

doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
We model it by incorporating the Cahn–Hilliard equation with a Turing mechanism of local activation/global inhibition
More here: www.nature.com/articles/s41...

We model it by incorporating the Cahn–Hilliard equation with a Turing mechanism of local activation/global inhibition
More here: www.nature.com/articles/s41...

www.biorxiv.org/cgi/content/...

www.biorxiv.org/cgi/content/...
biorxiv.org/content/10.110…
Postdoc @mewynne759 looked at microtubules, IFs/GFAP & actin along long elaborate processes.
Wonderful to collaborate w postdoc @dharshinigopal & @NogalesLab on cryo-ET 🤓
1/x
biorxiv.org/content/10.110…
Postdoc @mewynne759 looked at microtubules, IFs/GFAP & actin along long elaborate processes.
Wonderful to collaborate w postdoc @dharshinigopal & @NogalesLab on cryo-ET 🤓
1/x
www.biorxiv.org/content/10.1...
#teamtomo

www.biorxiv.org/content/10.1...
#teamtomo
Thankful to my mentors Kara Marshall and Blair Benham-Pyle for helping me chase this project through the last 1.5 years. 🏃♀️


Thankful to my mentors Kara Marshall and Blair Benham-Pyle for helping me chase this project through the last 1.5 years. 🏃♀️
www.nature.com/articles/s41...
www.nature.com/articles/s41...


(1) Accurate sequencing of sperm at scale
(2) Positive selection of spermatogenesis driver mutations across the exome
(3) Offspring disease risks from male reproductive aging
[1/n]
www.nature.com/articles/s41...

(1) Accurate sequencing of sperm at scale
(2) Positive selection of spermatogenesis driver mutations across the exome
(3) Offspring disease risks from male reproductive aging
[1/n]
www.nature.com/articles/s41...
Kampmeyer et al, 2023:
Lysine deserts prevent adventitious ubiquitylation of ubiquitin-proteasome components
doi.org/10.1007/s000...

Kampmeyer et al, 2023:
Lysine deserts prevent adventitious ubiquitylation of ubiquitin-proteasome components
doi.org/10.1007/s000...
Link to preprint: tinyurl.com/5npjkyps
#IDRs #condensates #CBP

Link to preprint: tinyurl.com/5npjkyps
#IDRs #condensates #CBP