
In her work as a medical researcher, she became a leading expert in histochemistry and enzyme kinetics while fighting against sexism in her field.
Let's learn more about her life!
🧵1/10

In her work as a medical researcher, she became a leading expert in histochemistry and enzyme kinetics while fighting against sexism in her field.
Let's learn more about her life!
🧵1/10
If you are looking for some advice about microprotein evidence, check out the latest from our consortium. Lead by @carvunis.bsky.social and Aaron Wacholder!
🧬 💻
#genomics
#RNASky
bit.ly/4bgJSDm

If you are looking for some advice about microprotein evidence, check out the latest from our consortium. Lead by @carvunis.bsky.social and Aaron Wacholder!
🧬 💻
#genomics
#RNASky
bit.ly/4bgJSDm
I rarely see the point in sharing disapproval for published papers. Let bygones be bygones, I suppose.
Several days ago, however, a new paper came out. My assessment is that it is bogus.
#genomics
🧬 💻
1/
www.nature.com/articles/s41...

I rarely see the point in sharing disapproval for published papers. Let bygones be bygones, I suppose.
Several days ago, however, a new paper came out. My assessment is that it is bogus.
#genomics
🧬 💻
1/
www.nature.com/articles/s41...
rdcu.be/d5vRC
rdcu.be/d5vRC
The RNAs evolved to regulate themselves in an appropriate way, taking advantage of a protein whose structure changed depending on the cellular iron status.
22/n

The RNAs evolved to regulate themselves in an appropriate way, taking advantage of a protein whose structure changed depending on the cellular iron status.
22/n


Eds: Stan Fields (UW Seattle), Maria Chikina (U Pittsburgh), Konrad Lohse (U Edinburgh).
Deadline Mar 1, 2025.
click.skem1.com/click/bbu3-2...

Eds: Stan Fields (UW Seattle), Maria Chikina (U Pittsburgh), Konrad Lohse (U Edinburgh).
Deadline Mar 1, 2025.
click.skem1.com/click/bbu3-2...
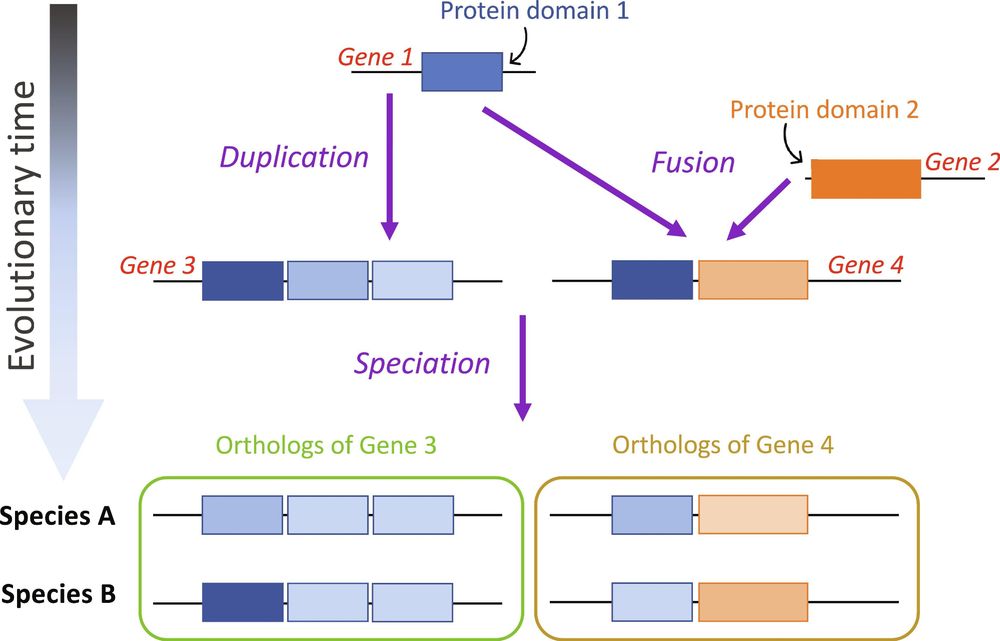

Our new preprint by Taylor Mighell
www.biorxiv.org/content/10.1...
thread on the other place: x.com/taylor_mighe...

Our new preprint by Taylor Mighell
www.biorxiv.org/content/10.1...
thread on the other place: x.com/taylor_mighe...


Check out microproteins!
Thanks to @science.org for featuring this work, in collaboration with @sebastiaanvheesch.bsky.social, #HUPO, @gencodegenes.bsky.social
www.science.org/content/arti...
🔎 microprotein
🧬 💻
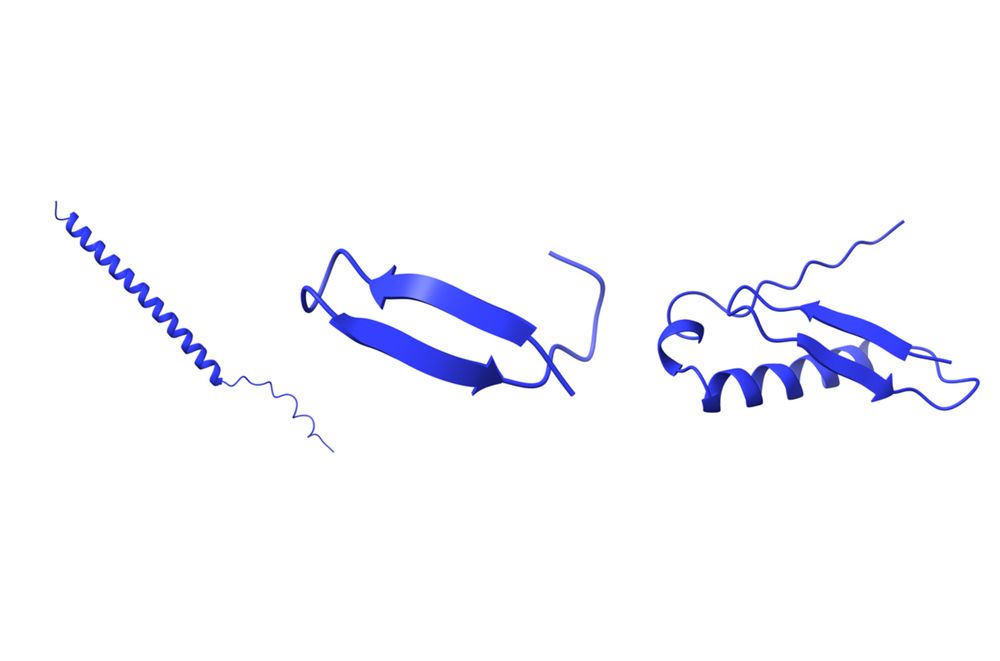
Check out microproteins!
Thanks to @science.org for featuring this work, in collaboration with @sebastiaanvheesch.bsky.social, #HUPO, @gencodegenes.bsky.social
www.science.org/content/arti...
🔎 microprotein
🧬 💻
www.biorxiv.org/content/10.1...
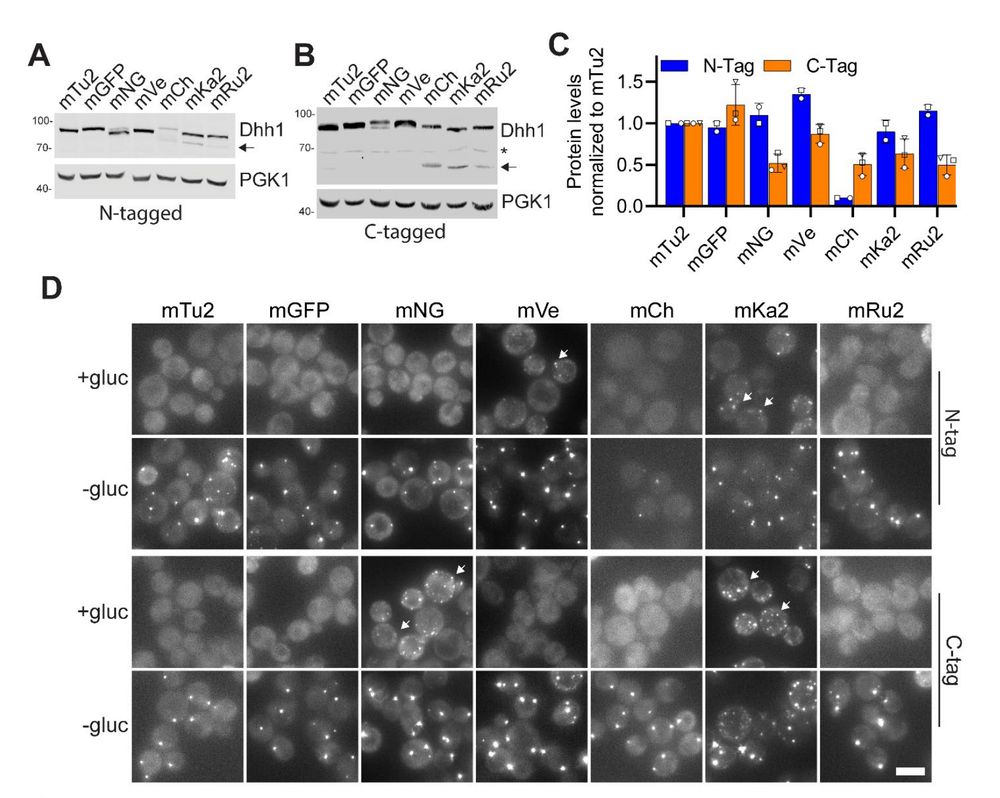
www.biorxiv.org/content/10.1...
We have Assistant and Associate Professor positions! 🚀
Pittsburgh is beautiful, and we are doing amazing science here.
cfopitt.taleo.net/careersectio...
cfopitt.taleo.net/careersectio...
Please apply by Dec 2!
So many wonderful advances in this area now make FFPE a rich source for study of RNA, DNA and protein.
Check out this new gem. 💎
#proteomics
#genomics
www.biorxiv.org/content/10.1...

So many wonderful advances in this area now make FFPE a rich source for study of RNA, DNA and protein.
Check out this new gem. 💎
#proteomics
#genomics
www.biorxiv.org/content/10.1...
@carvunis.bsky.social
@thomasmartinez.bsky.social
@sebastiaanvheesch.bsky.social
@microproteins.bsky.social
@nickywhiffin.bsky.social
@uweohler.bsky.social
@luispedrocoelho.bsky.social
@slavofflab.bsky.social

@carvunis.bsky.social
@thomasmartinez.bsky.social
@sebastiaanvheesch.bsky.social
@microproteins.bsky.social
@nickywhiffin.bsky.social
@uweohler.bsky.social
@luispedrocoelho.bsky.social
@slavofflab.bsky.social
---
#proteomics #prot-other

---
#proteomics #prot-other

They are stable & abundant O( 10³ ) copies / cell.
Generative mechanisms include codon-anticodon mismatches & RNA modifications.
Their abundance depends on codon frequency & protein stability.
www.biorxiv.org/content/10.1...

They are stable & abundant O( 10³ ) copies / cell.
Generative mechanisms include codon-anticodon mismatches & RNA modifications.
Their abundance depends on codon frequency & protein stability.
www.biorxiv.org/content/10.1...

