#HTS #Screening #ChemSky #DrugDiscovery

#HTS #Screening #ChemSky #DrugDiscovery
If you want to check it out, you can find it here: www.nature.com/articles/s41...
#ChemSky #ChemBio #ChemicalProteomics #Chemoproteomics #ProteoProbes #ChemPro

We have developed a new strategy termed "potency coherence analysis" that leverages the drug potency dimension in decryptM to decode the kinases that shape the human phosphoproteome.
Read more:

We have developed a new strategy termed "potency coherence analysis" that leverages the drug potency dimension in decryptM to decode the kinases that shape the human phosphoproteome.
Read more:
#ChemSky #ChemBio
#ChemSky #ChemBio
onlinelibrary.wiley.com/doi/10.1002/...
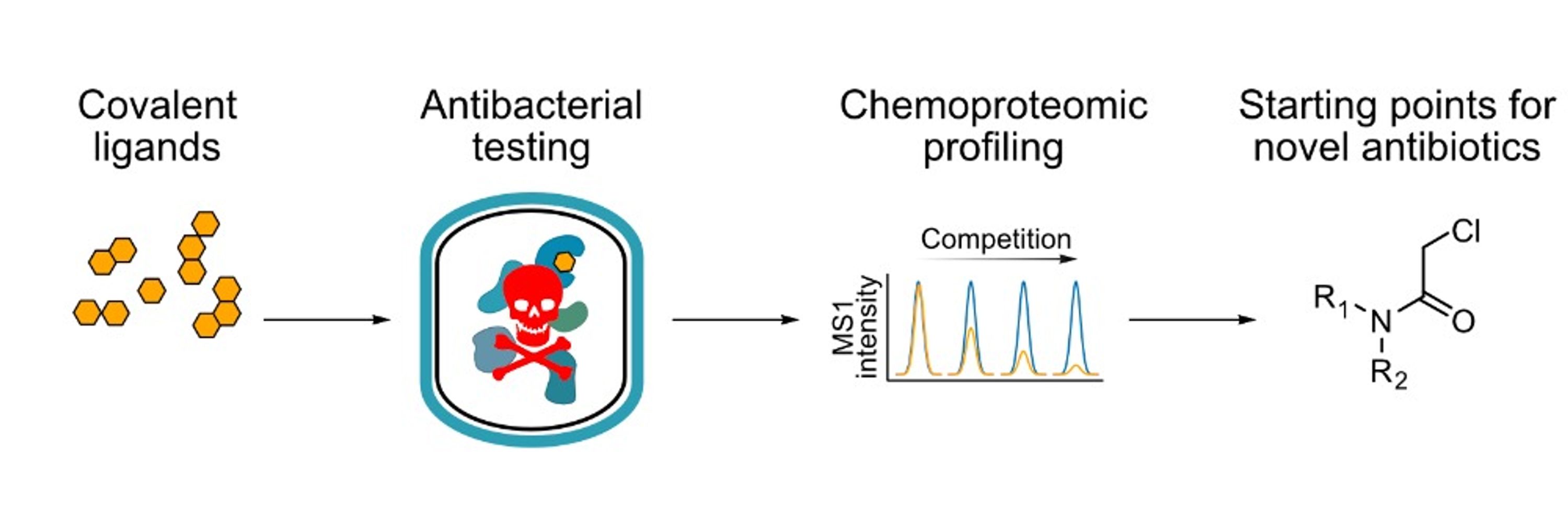
#Antibiotics #ChemSky
onlinelibrary.wiley.com/doi/10.1002/...
#Antibiotics #ChemSky
If you want to check it out, you can find it here: www.nature.com/articles/s41...
#ChemSky #ChemBio #ChemicalProteomics #Chemoproteomics #ProteoProbes #ChemPro

If you want to check it out, you can find it here: www.nature.com/articles/s41...
#ChemSky #ChemBio #ChemicalProteomics #Chemoproteomics #ProteoProbes #ChemPro
onlinelibrary.wiley.com/doi/10.1002/...
#ChemSky #ChemBio

onlinelibrary.wiley.com/doi/10.1002/...
#ChemSky #ChemBio
doi.org/10.1021/jacs...
#ChemSky #ChemBio #ChemicalProteomics #DrugDiscovery

doi.org/10.1021/jacs...
#ChemSky #ChemBio #ChemicalProteomics #DrugDiscovery
www.nature.com/articles/s41...
#ChemSky #ChemBio #ChemPro #ProteoProbes

www.nature.com/articles/s41...
#ChemSky #ChemBio #ChemPro #ProteoProbes
www.pnas.org/doi/10.1073/...
#ChemSky #ChemBio

www.pnas.org/doi/10.1073/...
#ChemSky #ChemBio
pubs.acs.org/doi/full/10....
#TPD #ChemSky #ChemBio #ChemicalProteomics

pubs.acs.org/doi/full/10....
#TPD #ChemSky #ChemBio #ChemicalProteomics
advanced.onlinelibrary.wiley.com/doi/10.1002/...
#TPD #ChemSky #ChemBio #DrugDiscovery #ChemicalProteomics #ChemPro #ProteoProbes
advanced.onlinelibrary.wiley.com/doi/10.1002/...
#TPD #ChemSky #ChemBio #DrugDiscovery #ChemicalProteomics #ChemPro #ProteoProbes
doi.org/10.1101/2025...

doi.org/10.1101/2025...
pubs.acs.org/doi/10.1021/...
#ChemSky #ChemBio #Peptides

pubs.acs.org/doi/10.1021/...
#ChemSky #ChemBio #Peptides
Our study on "Profiling the proteome-wide selectivity of diverse electrophiles" is published in Nature Chemistry.(1/7)
www.nature.com/articles/s41...

#ChemSky #ChemBio #DrugDiscovery
#ChemSky #ChemBio #DrugDiscovery
If you are interested, make sure to come by!
www.universiteitleiden.nl/en/science/l...
#ChemBio #ChemSky #DrugDiscovery

If you are interested, make sure to come by!
www.universiteitleiden.nl/en/science/l...
#ChemBio #ChemSky #DrugDiscovery
Next week, Lotte Bjerre Knudsen from Novo Nordisk will visit us to present on the development of GLP-1 receptor agonists.
If you are interested, make sure to come by.
#ChemBio #ChemSky #DrugDiscovery
If you are interested, make sure to come by!
www.universiteitleiden.nl/en/science/l...
#ChemBio #ChemSky #DrugDiscovery

Next week, Lotte Bjerre Knudsen from Novo Nordisk will visit us to present on the development of GLP-1 receptor agonists.
If you are interested, make sure to come by.
#ChemBio #ChemSky #DrugDiscovery
According to this new study by the @sebastianpomplun.bsky.social group of our LED3 hub, you may even get away without needing the DNA barcode. They describe self-encoded libraries (SELs) for #DrugDiscovery.
www.nature.com/articles/s41...
#ChemSky #ChemBio

According to this new study by the @sebastianpomplun.bsky.social group of our LED3 hub, you may even get away without needing the DNA barcode. They describe self-encoded libraries (SELs) for #DrugDiscovery.
www.nature.com/articles/s41...
#ChemSky #ChemBio
#TPD #ChemSky #ChemBio

#TPD #ChemSky #ChemBio
Check out the newest study by the group of @sebastianpomplun.bsky.social of our @led3hub.bsky.social. Being able to predict peptide binders solely based on structure may be closer than you think.
pubs.acs.org/doi/10.1021/...
#ChemSky #ChemBio

Check out the newest study by the group of @sebastianpomplun.bsky.social of our @led3hub.bsky.social. Being able to predict peptide binders solely based on structure may be closer than you think.
pubs.acs.org/doi/10.1021/...
#ChemSky #ChemBio
According to the newest paper by the group of @sebastianpomplun.bsky.social, the software tool #BindCraft allows you to predict such binders with high success rates.
pubs.acs.org/doi/10.1021/...
#ChemSky #ChemBio #DrugDiscovery

According to the newest paper by the group of @sebastianpomplun.bsky.social, the software tool #BindCraft allows you to predict such binders with high success rates.
pubs.acs.org/doi/10.1021/...
#ChemSky #ChemBio #DrugDiscovery
#ChemSky

#ChemSky
#ChemBio #ChemSky #DrugDiscovery



#ChemBio #ChemSky #DrugDiscovery
#ChemSky #ChemBio #DrugDiscovery #ABPP
www.nature.com/articles/s41...
#ChemSky #ChemBio #DrugDiscovery #ABPP
www.nature.com/articles/s41...

