Picture by Gatis Šļūka

Picture by Gatis Šļūka
Learn more or donate here:
www.dagstuhl.de/en/dblp/donate
Thank you very much!
Learn more or donate here:
www.dagstuhl.de/en/dblp/donate
Thank you very much!
k-mer indexes are the backbone of fast search in genomic data, but many degrade under small k, subsampling, or high diversity.
With Ondřej Sladký and @pavelvesely.bsky.social we asked: can we build one that works efficiently for any k-mer set?
Full article available: https://doi.org/10.1093/bioadv/vbaf290
Authors include: @pavelvesely.bsky.social, @brinda.eu
k-mer indexes are the backbone of fast search in genomic data, but many degrade under small k, subsampling, or high diversity.
With Ondřej Sladký and @pavelvesely.bsky.social we asked: can we build one that works efficiently for any k-mer set?
Full article available: https://doi.org/10.1093/bioadv/vbaf290
Authors include: @pavelvesely.bsky.social, @brinda.eu

k-mer indexes are the backbone of fast search in genomic data, but many degrade under small k, subsampling, or high diversity.
With Ondřej Sladký and @pavelvesely.bsky.social we asked: can we build one that works efficiently for any k-mer set?
Full article available: https://doi.org/10.1093/bioadv/vbaf290
Authors include: @pavelvesely.bsky.social, @brinda.eu

Full article available: https://doi.org/10.1093/bioadv/vbaf290
Authors include: @pavelvesely.bsky.social, @brinda.eu
Tohle jedno slovo nejlépe vystihuje počin jednoho z nově zvolených ústavních činitelů téhle země
Začít mandát tím, že sundáte ukrajinskou vlajku z budovy pěkně ilustruje to, o co mu jde. Nikoliv o zlepšení téhle země, ale jen o rozdmýchávání vášní
Co s tím? Já si koupil drona

Tohle jedno slovo nejlépe vystihuje počin jednoho z nově zvolených ústavních činitelů téhle země
Začít mandát tím, že sundáte ukrajinskou vlajku z budovy pěkně ilustruje to, o co mu jde. Nikoliv o zlepšení téhle země, ale jen o rozdmýchávání vášní
Co s tím? Já si koupil drona


*AllTheBacteria 661k, multiline fasta*
gzip (pigz): 751GB
zstandard --long: 641GB (30% original size)
*Single line fasta*
gzip (pigz): 700GB
zstandard --long: 232GB (10% original size)
*AllTheBacteria 661k, multiline fasta*
gzip (pigz): 751GB
zstandard --long: 641GB (30% original size)
*Single line fasta*
gzip (pigz): 700GB
zstandard --long: 232GB (10% original size)
Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...

Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...
Modern Minimal Perfect Hashing: A Survey
https://arxiv.org/abs/2506.06536
11011110.github.io/blog/2025/05... via @theory.report
11011110.github.io/blog/2025/05... via @theory.report



Excitement is building for #RECOMBseq 2025 in Seoul 🎉
Join leading researchers as we dive into cutting-edge computational genomics, from single-cell to long-read sequencing.
🗓 April 24-25
📍 Seoul
📄 Program: recomb-seq.github.io/program/
#RECOMB2025 #Genomics #Bioinformatics
Excitement is building for #RECOMBseq 2025 in Seoul 🎉
Join leading researchers as we dive into cutting-edge computational genomics, from single-cell to long-read sequencing.
🗓 April 24-25
📍 Seoul
📄 Program: recomb-seq.github.io/program/
#RECOMB2025 #Genomics #Bioinformatics
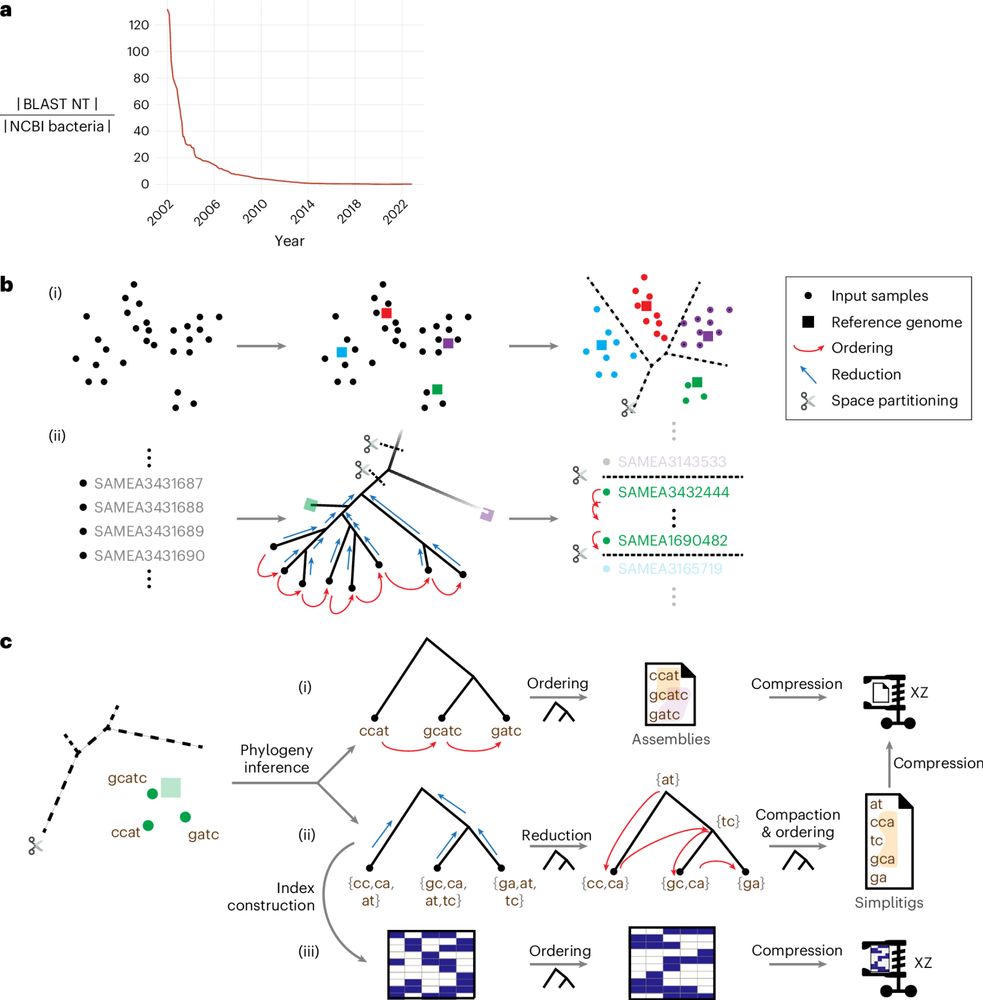
Our paper in @naturemethods.bsky.social answers this: use evolutionary history to guide compression and search.
rdcu.be/eg4OA
w/ @baym.lol, @zaminiqbal.bsky.social et al. 🧵1/

Our paper in @naturemethods.bsky.social answers this: use evolutionary history to guide compression and search.
rdcu.be/eg4OA
w/ @baym.lol, @zaminiqbal.bsky.social et al. 🧵1/
rdcu.be/eg4OA
1/