Full article available: https://doi.org/10.1093/bioadv/vbaf281

Full article available: https://doi.org/10.1093/bioadv/vbaf281
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf286
Authors include: @rnaprof.bsky.social

Read the full paper here: https://doi.org/10.1093/bioadv/vbaf286
Authors include: @rnaprof.bsky.social
Explore the full study: https://doi.org/10.1093/bioadv/vbaf280

Explore the full study: https://doi.org/10.1093/bioadv/vbaf280
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf287

Read the full paper here: https://doi.org/10.1093/bioadv/vbaf287
Full article available: https://doi.org/10.1093/bioadv/vbaf284

Full article available: https://doi.org/10.1093/bioadv/vbaf284
Read the full paper here:
Read the full paper here:
Full article available: https://doi.org/10.1093/bioadv/vbaf275
Authors include: @genomesevolve.bsky.social, @songfeng.bsky.social

Full article available: https://doi.org/10.1093/bioadv/vbaf275
Authors include: @genomesevolve.bsky.social, @songfeng.bsky.social
Explore the full study: https://doi.org/10.1093/bioadv/vbaf279

Explore the full study: https://doi.org/10.1093/bioadv/vbaf279
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf278

Read the full paper here: https://doi.org/10.1093/bioadv/vbaf278
Explore the full study: https://doi.org/10.1093/bioadv/vbaf255

Explore the full study: https://doi.org/10.1093/bioadv/vbaf255
Explore the full study: https://doi.org/10.1093/bioadv/vbaf276

Explore the full study: https://doi.org/10.1093/bioadv/vbaf276
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf277

Read the full paper here: https://doi.org/10.1093/bioadv/vbaf277
Full article available: https://doi.org/10.1093/bioadv/vbaf273
Authors include: @fvillanelo.bsky.social
Full article available: https://doi.org/10.1093/bioadv/vbaf273
Authors include: @fvillanelo.bsky.social
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf274
Authors include: @taanec.bsky.social

Read the full paper here: https://doi.org/10.1093/bioadv/vbaf274
Authors include: @taanec.bsky.social
Explore the full study: https://doi.org/10.1093/bioadv/vbaf271

Explore the full study: https://doi.org/10.1093/bioadv/vbaf271
Full article available: https://doi.org/10.1093/bioadv/vbaf272

Full article available: https://doi.org/10.1093/bioadv/vbaf272
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf263

Read the full paper here: https://doi.org/10.1093/bioadv/vbaf263
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf270
Authors include: @wachtenlab.bsky.social, @ecsignalmetabolism.bsky.social

Read the full paper here: https://doi.org/10.1093/bioadv/vbaf270
Authors include: @wachtenlab.bsky.social, @ecsignalmetabolism.bsky.social
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf269

Read the full paper here: https://doi.org/10.1093/bioadv/vbaf269
Explore the full study: https://doi.org/10.1093/bioadv/vbaf262
Authors include: @joellepasman.bsky.social

Explore the full study: https://doi.org/10.1093/bioadv/vbaf262
Authors include: @joellepasman.bsky.social
Read the full paper here: ttps://doi.org/10.1093/bioadv/vbaf268

Read the full paper here: ttps://doi.org/10.1093/bioadv/vbaf268
Full article available: https://doi.org/10.1093/bioadv/vbaf265
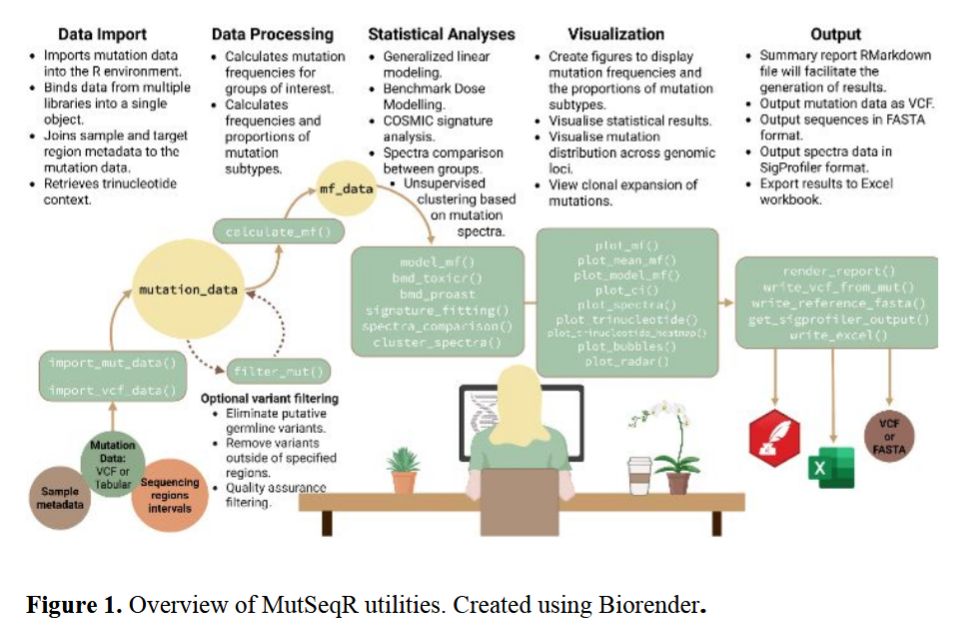
Full article available: https://doi.org/10.1093/bioadv/vbaf265
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf264
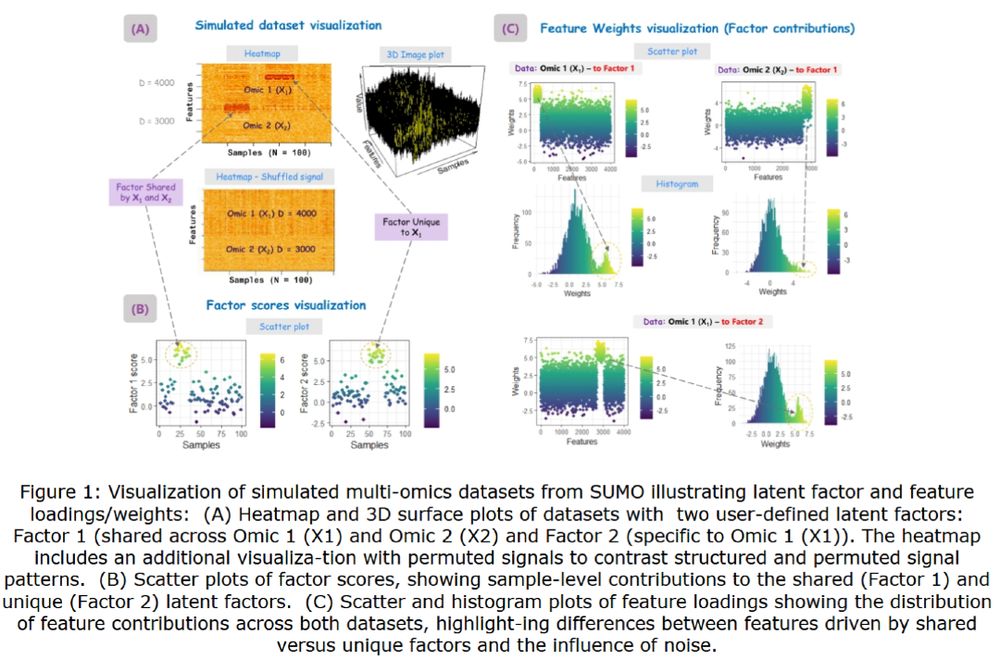
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf264
Explore the full study: https://doi.org/10.1093/bioadv/vbaf266

Explore the full study: https://doi.org/10.1093/bioadv/vbaf266
Full article available: https://doi.org/10.1093/bioadv/vbaf261

Full article available: https://doi.org/10.1093/bioadv/vbaf261

