
https://nikhilmilind.dev/
@simonsfoundation.org. Details to follow. Please RT.
@simonsfoundation.org. Details to follow. Please RT.
Amanda, a fantastic PhD student with Dan, led this "phylo-functional" work & uncovered complex duplication & loss of a glucosinolate gene across the Brassicaceae affecting enantiomeric specificity
Amanda, a fantastic PhD student with Dan, led this "phylo-functional" work & uncovered complex duplication & loss of a glucosinolate gene across the Brassicaceae affecting enantiomeric specificity
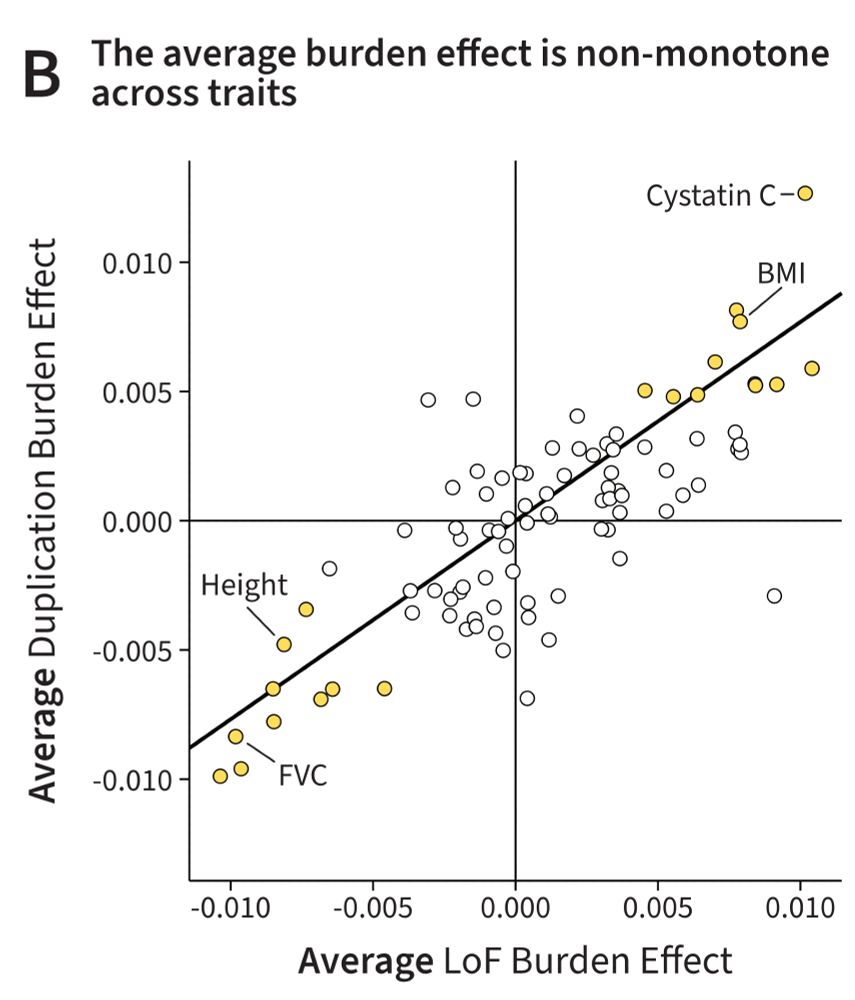
In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...

We describe a critical concept that we call *specificity*.
Led by Jeff Spence and Hakhamanesh Mostafavi:
In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...

We describe a critical concept that we call *specificity*.
Led by Jeff Spence and Hakhamanesh Mostafavi:
More fundamentally, how SHOULD we rank genes?
We propose two possible criteria: trait IMPORTANCE and trait SPECIFICITY.
5/n
More fundamentally, how SHOULD we rank genes?
We propose two possible criteria: trait IMPORTANCE and trait SPECIFICITY.
5/n
In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...

In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...

In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...

In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...
academicjobsonline.org/ajo/jobs/30328
academicjobsonline.org/ajo/jobs/30328
In our new PLOS Biology paper, we will try to convince you that two simple scaling laws drive differences in the number, effect sizes and frequencies of causal variants affecting complex traits.
Thread:
journals.plos.org/plosbiology/...

In our new PLOS Biology paper, we will try to convince you that two simple scaling laws drive differences in the number, effect sizes and frequencies of causal variants affecting complex traits.
Thread:
journals.plos.org/plosbiology/...
Registration is free but required. The deadline for talk submission is Nov. 16. Hope to see you soon! Pls RT!
docs.google.com/forms/d/e/1F...
Registration is free but required. The deadline for talk submission is Nov. 16. Hope to see you soon! Pls RT!
docs.google.com/forms/d/e/1F...
It’s not your imagination. It’s climate change.
Published in the November issue of Journal of the Atmospheric Sciences.
doi.org/10.1175/JAS-...

It’s not your imagination. It’s climate change.
👉 I'll be presenting a poster on our new work on genome-wide perturb-seq screens in primary human T cells (5049W, Wed 2.30pm)
👉 you can hear me talk about it at the Industry Education session presented by Ultima Genomics (Thu 3pm) [1/2]
I did this last year and it was great to meet a whole bunch of new people, at all career stages!
I did this last year and it was great to meet a whole bunch of new people, at all career stages!
www.medrxiv.org/content/10.1...
www.medrxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
(1/n)

www.biorxiv.org/content/10.1...
(1/n)
doi.org/10.1038/s415...
It was a long journey – 16 months from initial submission to acceptance. Is it just me, or has peer review gotten more arduous lately? 4+ rounds of review isn't so unusual these days...

doi.org/10.1038/s415...
It was a long journey – 16 months from initial submission to acceptance. Is it just me, or has peer review gotten more arduous lately? 4+ rounds of review isn't so unusual these days...
The expected answer: selection.
But our new paper in GENETICS shows that genetic drift alone can generate sexual dimorphism — even when male & female optima are the same
The expected answer: selection.
But our new paper in GENETICS shows that genetic drift alone can generate sexual dimorphism — even when male & female optima are the same
We show how data on expression quantitative trait loci (eQTL) relates to the structure of gene regulatory networks (GRN). Much of the GRN / eQTL picture is unmapped, but what we do have says a lot… (1/)
doi.org/10.1101/2025...
We show how data on expression quantitative trait loci (eQTL) relates to the structure of gene regulatory networks (GRN). Much of the GRN / eQTL picture is unmapped, but what we do have says a lot… (1/)
doi.org/10.1101/2025...



