@haesleinhuepf.bsky.social
for image registration purposes I need to select matching landmarks from images.
Are you aware of a jupyter widget that can help with that?
I don't want to download all my images from my HPC.
@haesleinhuepf.bsky.social
for image registration purposes I need to select matching landmarks from images.
Are you aware of a jupyter widget that can help with that?
I don't want to download all my images from my HPC.
there is interest in showcasing STalign in a big spatial tutorial.
However the current requirements.txt make it difficult to include.
Could you take a look at
github.com/JEFworks-Lab...
there is interest in showcasing STalign in a big spatial tutorial.
However the current requirements.txt make it difficult to include.
Could you take a look at
github.com/JEFworks-Lab...
https://www.biorxiv.org/content/10.1101/2025.03.31.646342v1
https://www.biorxiv.org/content/10.1101/2025.03.31.646342v1
If so, you might this little project: github.com/rrwick/conda...
If so, you might this little project: github.com/rrwick/conda...
Keep reading for more about how we did the study and what we found out 🧵 👇
1/16
Keep reading for more about how we did the study and what we found out 🧵 👇
1/16

please enjoy all analyses equally
👉 lucymcgowan.github.io/mdr-website/
and for my #rstats friends:
👉 lucymcgowan.github.io/mdr/
please enjoy all analyses equally
👉 lucymcgowan.github.io/mdr-website/
and for my #rstats friends:
👉 lucymcgowan.github.io/mdr/
@brukerspatial.bsky.social
In www.nature.com/articles/s41...
You presented a dataset with 980 RNAs and 108 proteins.
But on staging.nanostring.com/products/cos... I only find the RNA and a couple of protein markers for QC and segmentation.
Where can I find the rest of the protein data?
Ty!
@brukerspatial.bsky.social
In www.nature.com/articles/s41...
You presented a dataset with 980 RNAs and 108 proteins.
But on staging.nanostring.com/products/cos... I only find the RNA and a couple of protein markers for QC and segmentation.
Where can I find the rest of the protein data?
Ty!
Here is how and why I gave this model organism a visual upgrade 🧵(1/7)

Here is how and why I gave this model organism a visual upgrade 🧵(1/7)
nbsanity.com
It now works with gists!

nbsanity.com
It now works with gists!

💡 idea is to extend mutual nearest neighbors for
#spatial data. We call it spatial mutual nearest neighbors (spatialMNN) 😄
Thank you @haowen-zhou.bsky.social @pratibha-panwar.bsky.social who led this work! 👏 🧬🖥️🧪
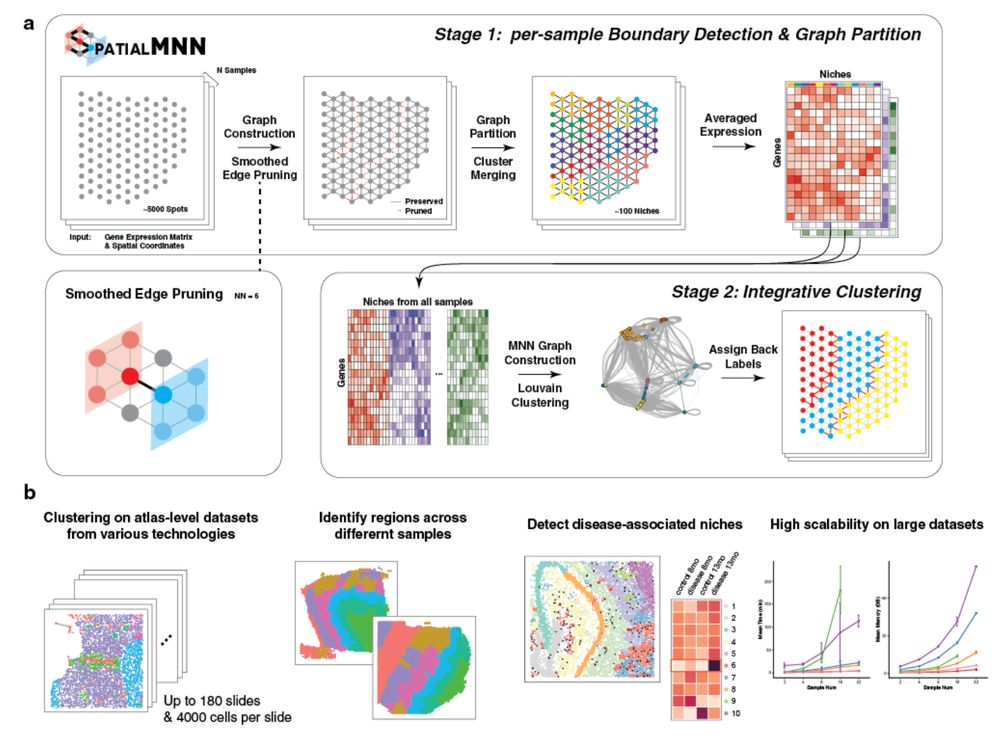
💡 idea is to extend mutual nearest neighbors for
#spatial data. We call it spatial mutual nearest neighbors (spatialMNN) 😄
Thank you @haowen-zhou.bsky.social @pratibha-panwar.bsky.social who led this work! 👏 🧬🖥️🧪


swearsky.bagpuss.org

swearsky.bagpuss.org
We use spectral graph theory to extract 1D curves related to tissue morphology, creating a more relevant coordinate system. Improves sensitivity.
with @PhillipNicol et al.
www.biorxiv.org/content/10.1...
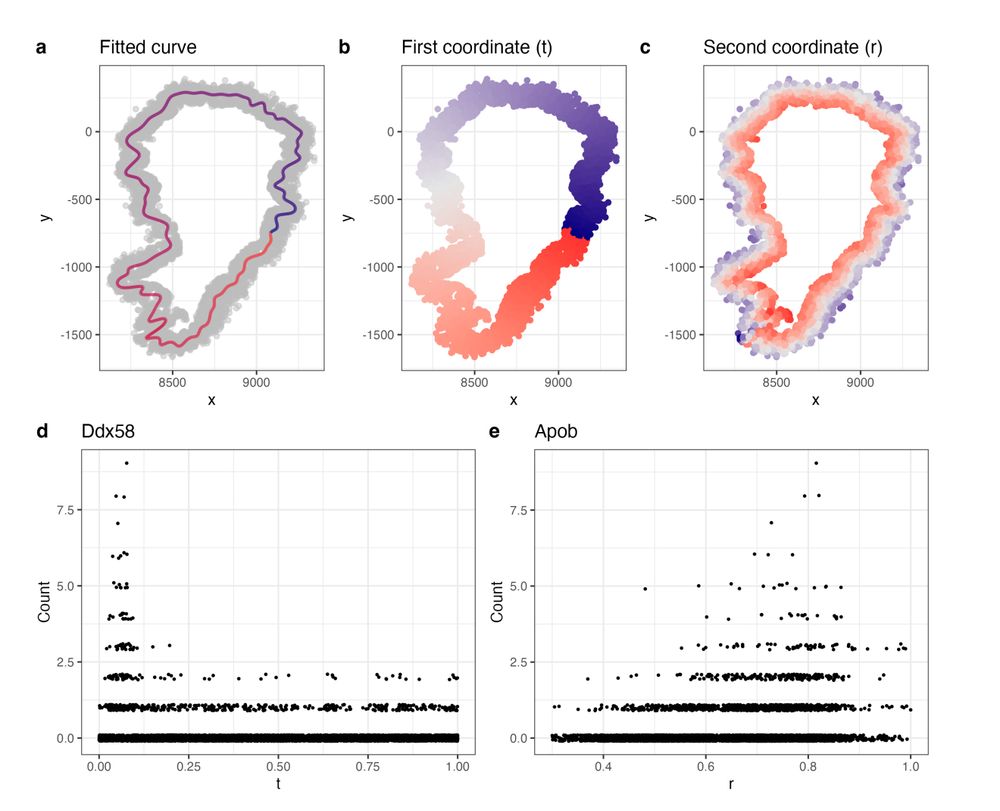
We use spectral graph theory to extract 1D curves related to tissue morphology, creating a more relevant coordinate system. Improves sensitivity.
with @PhillipNicol et al.
www.biorxiv.org/content/10.1...
www.youtube.com/playlist?lis...
I usually have my interns watch these videos to get introduced to the basic concepts before I go show them how to in the lab.
www.youtube.com/playlist?lis...
I usually have my interns watch these videos to get introduced to the basic concepts before I go show them how to in the lab.
Starter packs for genomics, bioinformatics, #Rstats, Nextflow. Moderation lists. Feeds. Let's rebuild the old scitwitter community and keep this place nice
blog.stephenturner.us/p/bluesky-fo... 🧬🖥️🧪

Starter packs for genomics, bioinformatics, #Rstats, Nextflow. Moderation lists. Feeds. Let's rebuild the old scitwitter community and keep this place nice
blog.stephenturner.us/p/bluesky-fo... 🧬🖥️🧪

@prepub-singlecell.bsky.social
(I posted about it a little while ago when no one was here, so posting again now :D)
It's pretty dumb so very open to suggestions for improvements!
@prepub-singlecell.bsky.social
(I posted about it a little while ago when no one was here, so posting again now :D)
It's pretty dumb so very open to suggestions for improvements!

