🔹Join a research group in our department
🔹Co-mentoring opportunities with 2+ faculty
🔹Collaborate with investigators beyond our department
🔹Salary starts at $75K
Apply here: t.co/B7SLZzQFKu
🔹Join a research group in our department
🔹Co-mentoring opportunities with 2+ faculty
🔹Collaborate with investigators beyond our department
🔹Salary starts at $75K
Apply here: t.co/B7SLZzQFKu
Join us at @HarvardBiostats and @dfcidatascience
We are filling THREE positions:
1️⃣ Tenure-track (any rank) in AI/ML
2️⃣ Assistant Professor in Single Cell Genomics
3️⃣ Lecturer & Director Training/Education
Need to apply separately for each role. Links 👇

Join us at @HarvardBiostats and @dfcidatascience
We are filling THREE positions:
1️⃣ Tenure-track (any rank) in AI/ML
2️⃣ Assistant Professor in Single Cell Genomics
3️⃣ Lecturer & Director Training/Education
Need to apply separately for each role. Links 👇
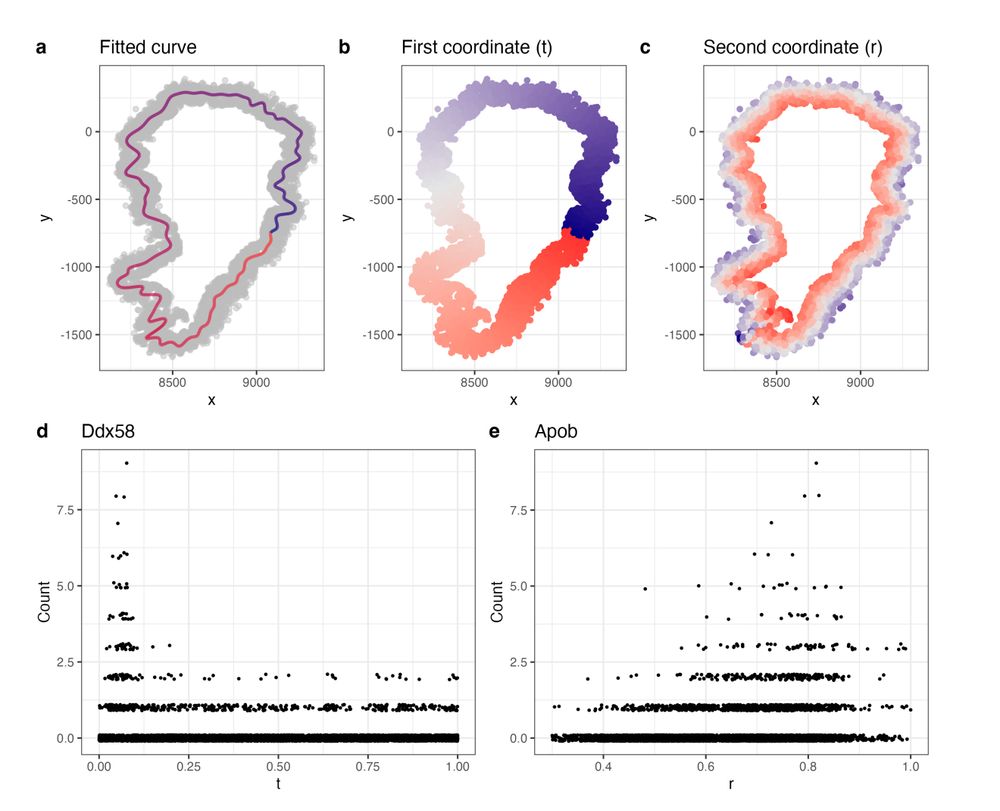
We use spectral graph theory to extract 1D curves related to tissue morphology, creating a more relevant coordinate system. Improves sensitivity.
with @PhillipNicol et al.
www.biorxiv.org/content/10.1...
We use spectral graph theory to extract 1D curves related to tissue morphology, creating a more relevant coordinate system. Improves sensitivity.
with @PhillipNicol et al.
www.biorxiv.org/content/10.1...
I have 100s of exercises along with solutions in quarto documents (see image). But they are not formatted in a systematic way. I would like to easily create docs with and w/o solutions and export to LMS.

I have 100s of exercises along with solutions in quarto documents (see image). But they are not formatted in a systematic way. I would like to easily create docs with and w/o solutions and export to LMS.
t.co/TGID4IwTSQ
t.co/TGID4IwTSQ
1) Communicating data insights.
2) Guiding analysis and spotting data issues.
While modern courses and books focus on the first, it's the second - exploratory data analysis - that is more important and harder to teach.
1) Communicating data insights.
2) Guiding analysis and spotting data issues.
While modern courses and books focus on the first, it's the second - exploratory data analysis - that is more important and harder to teach.
- Join a research group in our department
- Co-mentoring opportunities with 2+ faculty
- Collaborate with DFCI investigators beyond our department
- Salary starts at $75K
Apply here: ds.dfci.harvard.edu/postdocs/
- Join a research group in our department
- Co-mentoring opportunities with 2+ faculty
- Collaborate with DFCI investigators beyond our department
- Salary starts at $75K
Apply here: ds.dfci.harvard.edu/postdocs/

