
hilbertlab.org
Grüße vom Internationalen Zebrafisch & Medaka Kurs
👋🐟
Photo from teaching this morning for this week’s November German language edition. This FELASA-certificate course is offered via @ezrc.bsky.social at @kit.edu

Grüße vom Internationalen Zebrafisch & Medaka Kurs
👋🐟
Photo from teaching this morning for this week’s November German language edition. This FELASA-certificate course is offered via @ezrc.bsky.social at @kit.edu
Integrative Approaches for DNA Sequence-Controlled Functional Materials
We jointly cover scale-bridging simulation, machine learning & experiments from several groups.
advanced.onlinelibrary.wiley.com/doi/10.1002/...

Integrative Approaches for DNA Sequence-Controlled Functional Materials
We jointly cover scale-bridging simulation, machine learning & experiments from several groups.
advanced.onlinelibrary.wiley.com/doi/10.1002/...






Some travel impressions, including fancy and very white “Le Studium” on Strasbourg Central Campus 🤍
And, Gyoza 🥟




Some travel impressions, including fancy and very white “Le Studium” on Strasbourg Central Campus 🤍
And, Gyoza 🥟

Based in the group of our collaborator @vasilyzaburdaev.bsky.social Tim decisively contributed to at least 3 of our own lab’s foundational projects, which he showed today at “Max Planck Zentrum Physik und Medizin”
Some impressions




Based in the group of our collaborator @vasilyzaburdaev.bsky.social Tim decisively contributed to at least 3 of our own lab’s foundational projects, which he showed today at “Max Planck Zentrum Physik und Medizin”
Some impressions
Mona Wellhäuser, MC15 @mofrawe.bsky.social
Yuzhi Bao, CA03 @yuzhibao.bsky.social
Lennart Hilbert, MC06
Conference abstracts:
gbm-compact.org/abstracts.html
*sketch added as click bait

Mona Wellhäuser, MC15 @mofrawe.bsky.social
Yuzhi Bao, CA03 @yuzhibao.bsky.social
Lennart Hilbert, MC06
Conference abstracts:
gbm-compact.org/abstracts.html
*sketch added as click bait
Read more about it here:
nyaspubs.onlinelibrary.wiley.com/doi/10.1111/...
Read more about it here:
nyaspubs.onlinelibrary.wiley.com/doi/10.1111/...





Highly conceptual Perspective article, supported by fresh data & simulations from our group. Containing my now 40 years of accumulated "wisdom".
nyaspubs.onlinelibrary.wiley.com/doi/10.1111/...

Highly conceptual Perspective article, supported by fresh data & simulations from our group. Containing my now 40 years of accumulated "wisdom".
nyaspubs.onlinelibrary.wiley.com/doi/10.1111/...




Gotta love these classic papers about nuclear architecture. In hindsight, all that we understand and discover today was already contained in their images.
www.sciencedirect.com/science/arti...

Gotta love these classic papers about nuclear architecture. In hindsight, all that we understand and discover today was already contained in their images.
www.sciencedirect.com/science/arti...
"Increased enhancer–promoter interactions during developmental enhancer activation in mammals"
www.nature.com/articles/s41...
@evgenykvon.bsky.social


"Increased enhancer–promoter interactions during developmental enhancer activation in mammals"
www.nature.com/articles/s41...
@evgenykvon.bsky.social
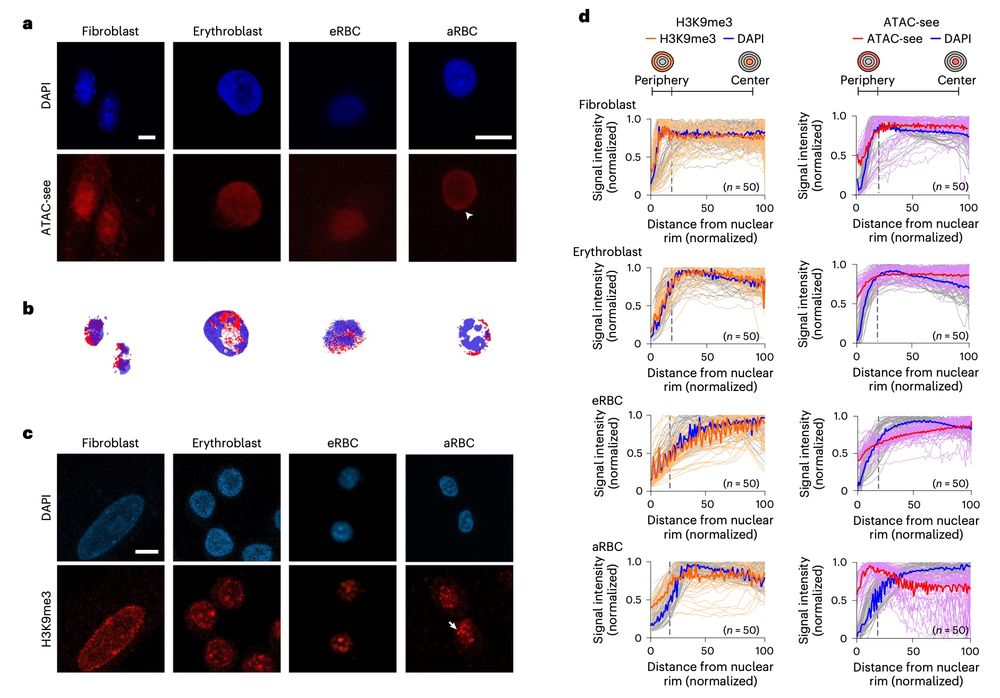
Excellent paper recommendation from @mariekeoudelaar.bsky.social Thanks!
Andrés Penagos-Puig, ... Mayra Furlan-Magaril
www.nature.com/articles/s41...


Excellent paper recommendation from @mariekeoudelaar.bsky.social Thanks!
Andrés Penagos-Puig, ... Mayra Furlan-Magaril
www.nature.com/articles/s41...
Our PhD student @mofrawe.bsky.social learned the protocol from amazing @mpownall.bsky.social 🙏
Months of preparation are paying off now!

Our PhD student @mofrawe.bsky.social learned the protocol from amazing @mpownall.bsky.social 🙏
Months of preparation are paying off now!


www.pse.kit.edu/english/karr...
www.pse.kit.edu/english/karr...

