




lancasteronline.com/opinion/colu...
lancasteronline.com/opinion/colu...

This paper was lead by @niraj-rayamajhi.bsky.social, in collaboration with Bushra Fazal Minhas, Chris Cheng, and @jcatchen.bsky.social.
doi.org/10.1093/g3jo...
1/n
In a new preprint
www.biorxiv.org/content/10.1...
we merge single-cell RNAseq (defining stickleback cell types) and population genomic measures of selection...

In a new preprint
www.biorxiv.org/content/10.1...
we merge single-cell RNAseq (defining stickleback cell types) and population genomic measures of selection...
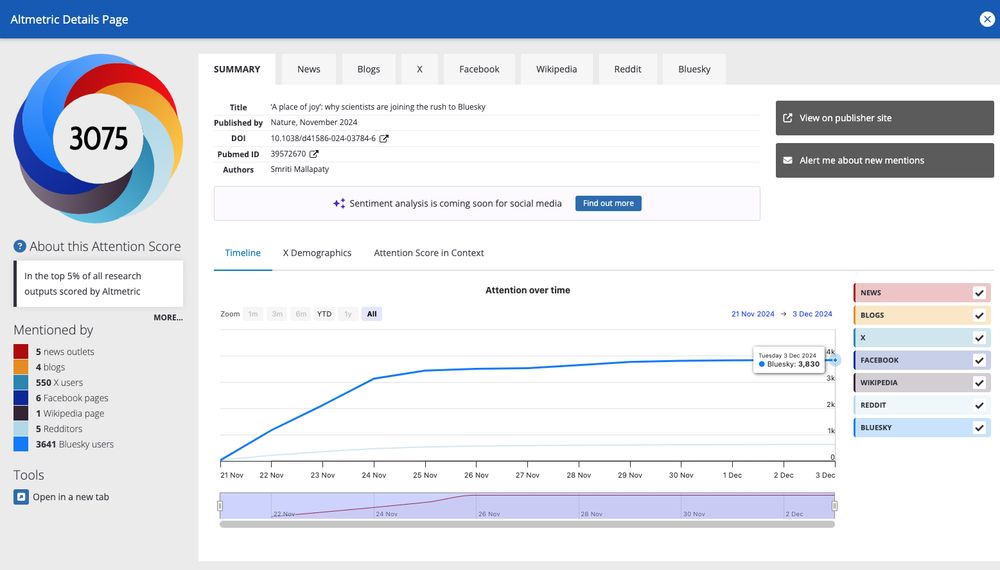
A Place of Joy.
We've been adding over a million users per day for the last few days. To celebrate, here are 20 fun facts about Bluesky:
We've been adding over a million users per day for the last few days. To celebrate, here are 20 fun facts about Bluesky:
This includes funds for a postdoc and PhD student! I can be reached at miltont (at) illinois (dot) edu. 🐟





catchenlab.life.illinois.edu/stacks/
catchenlab.life.illinois.edu/stacks/


"GWAS variants differ from eQTL variants along many axes…GWAS hits are biased toward more constrained genes…with complex regulatory landscapes…eQTLs show strong promoter bias largely absent from GWAS hits"

"GWAS variants differ from eQTL variants along many axes…GWAS hits are biased toward more constrained genes…with complex regulatory landscapes…eQTLs show strong promoter bias largely absent from GWAS hits"
And how white-blooded #icefish lost their hemoglobin genes?
Our new MS is for you!
A quick 🧵on our finding in @MolBioEvol!
#Marinelife
tinyurl.com/5n7ym3jp

And how white-blooded #icefish lost their hemoglobin genes?
Our new MS is for you!
A quick 🧵on our finding in @MolBioEvol!
#Marinelife
tinyurl.com/5n7ym3jp
The Practice of RADseq: Population Genomics Analysis with Stacks, October 2-6, 2023
The course will focus on de novo (with parameter optimization) and reference analyses of RAD data, with new modules on PCR duplicates/allele dropout and private allele analysis

The Practice of RADseq: Population Genomics Analysis with Stacks, October 2-6, 2023
The course will focus on de novo (with parameter optimization) and reference analyses of RAD data, with new modules on PCR duplicates/allele dropout and private allele analysis



