I am excited to announce that our new study explaining the missing heritability of many phenotypes using WGS data from ~347,000 UK Biobank participants has just been published in @Nature.
Our manuscript is here: www.nature.com/articles/s41....

I am excited to announce that our new study explaining the missing heritability of many phenotypes using WGS data from ~347,000 UK Biobank participants has just been published in @Nature.
Our manuscript is here: www.nature.com/articles/s41....
We’ll be providing our filtered AoU WGS plink pgens for all registered users: watch this space
We show why you should do genotype-level QC on your WGS data
www.biorxiv.org/content/10.1...
Very real quotes about this paper -
“The most exciting, mind-blowing paper of the year!”
“On a par with Fisher 1918”
“I read it every night. Just so beautiful”

We’ll be providing our filtered AoU WGS plink pgens for all registered users: watch this space
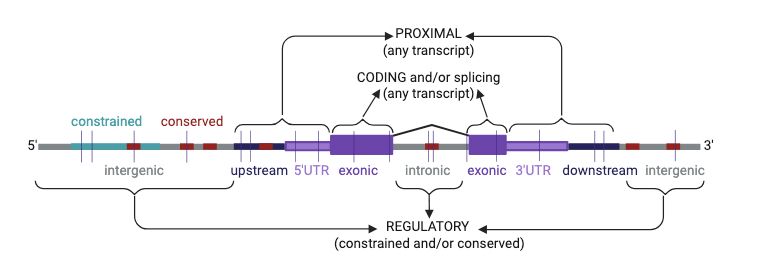
Come join if you are interested in de novo mutations, non-coding genome, rare disease, or just up for seeing @nickywhiffin.bsky.social talk about RNU4-2 saturation mutagenesis after me 😂
#ESHG2025
Come join if you are interested in de novo mutations, non-coding genome, rare disease, or just up for seeing @nickywhiffin.bsky.social talk about RNU4-2 saturation mutagenesis after me 😂
#ESHG2025

While this seems at odds with our recent work about how burden tests and GWAS prioritize different genes, our results agree (🧬🧪🧵 1/6)
www.biorxiv.org/content/10.1...

While this seems at odds with our recent work about how burden tests and GWAS prioritize different genes, our results agree (🧬🧪🧵 1/6)
www.biorxiv.org/content/10.1...
tinyurl.com/3wdfc2me
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
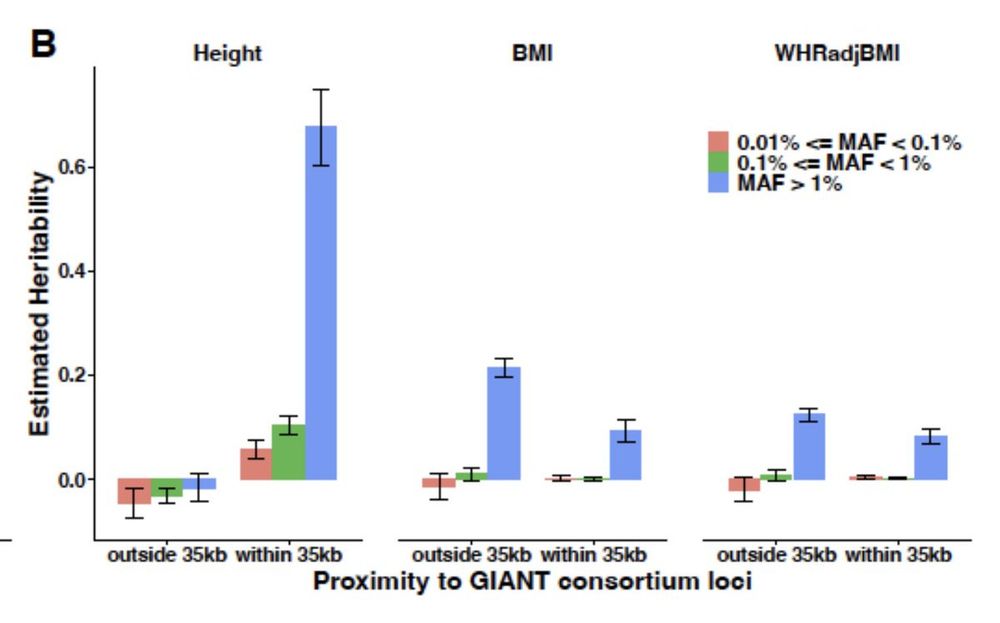
www.biorxiv.org/content/10.1...
Whole-genome sequencing analysis of anthropometric traits in 672,976 individuals reveals convergence between rare and common genetic associations www.biorxiv.org/content/10.1...

Whole-genome sequencing analysis of anthropometric traits in 672,976 individuals reveals convergence between rare and common genetic associations www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
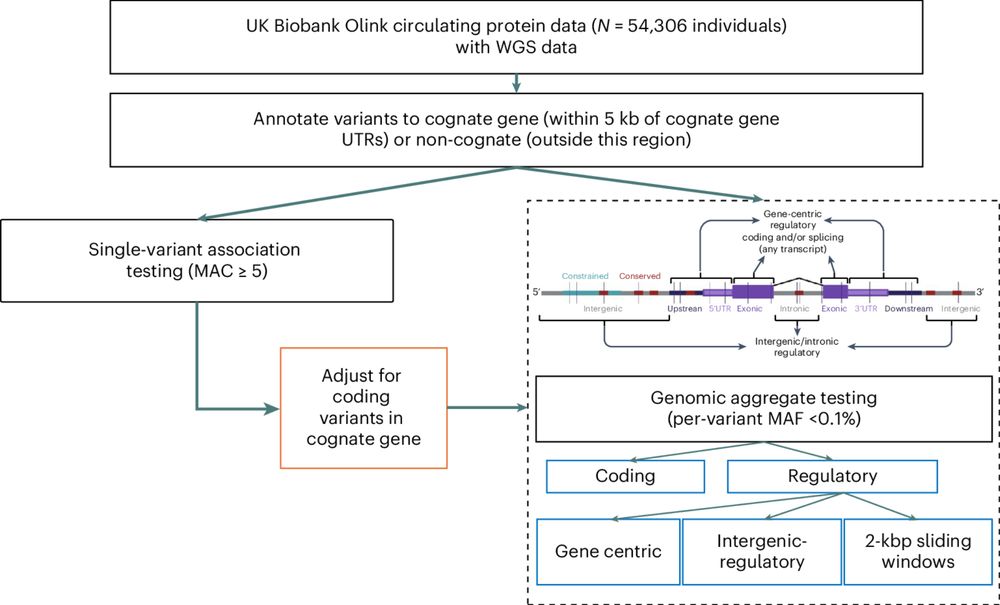
tinyurl.com/3wdfc2me
tinyurl.com/3wdfc2me



