
go.nature.com/48uEnAn

go.nature.com/48uEnAn
www.science.org/doi/10.1126/... #TBsky

www.science.org/doi/10.1126/... #TBsky


Today in @Nature, we share semantic design—a strategy for function-guided design with genomic language models that leverages genomic context to create de novo genes and systems with desired functions. 🧵
www.nature.com/articles/s41...
Today in @Nature, we share semantic design—a strategy for function-guided design with genomic language models that leverages genomic context to create de novo genes and systems with desired functions. 🧵
www.nature.com/articles/s41...
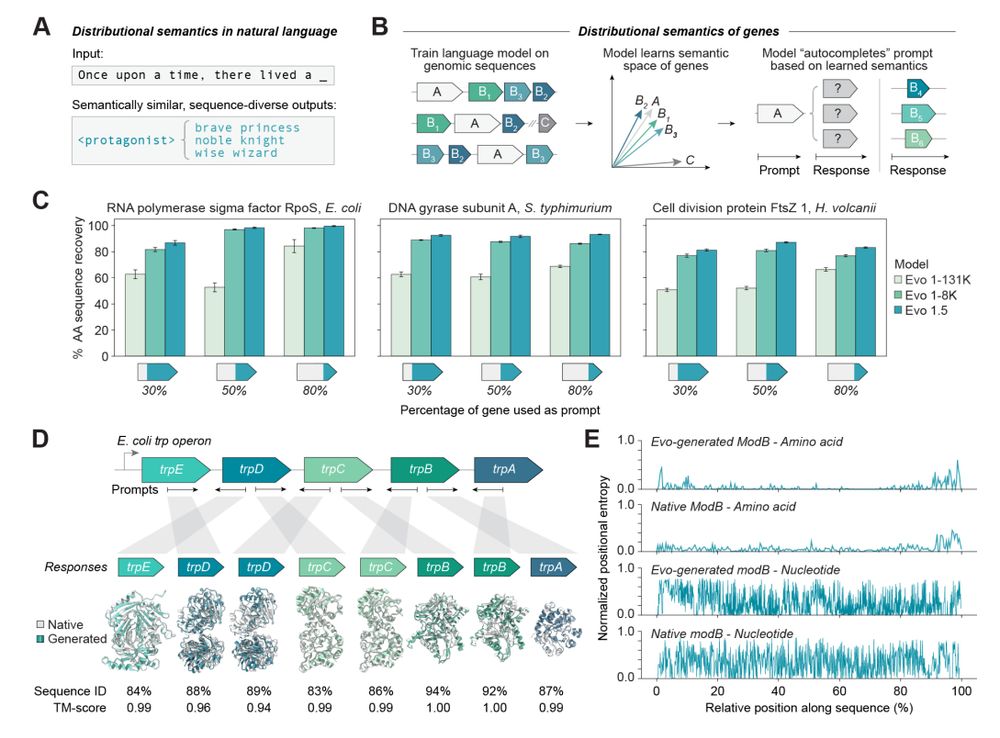
You shall know a gene by the company it keeps!

You shall know a gene by the company it keeps!
@synbiogaolab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...



@synbiogaolab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...

Germinal produces functional nanobodies in just dozens of tests, making custom antibody design more accessible than ever before.

Germinal produces functional nanobodies in just dozens of tests, making custom antibody design more accessible than ever before.
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...


@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
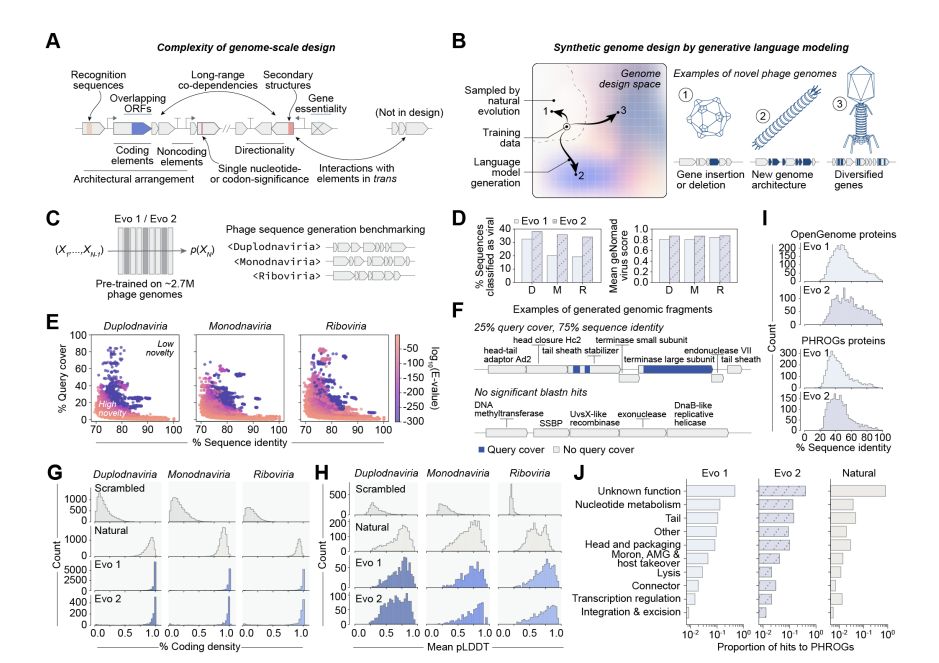
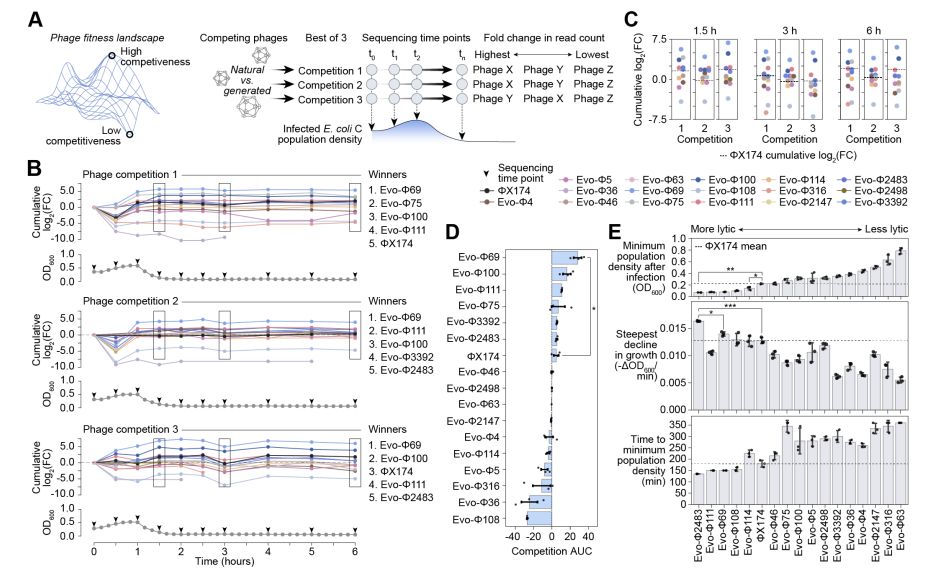
Generative design of novel bacteriophages with genome language models www.biorxiv.org/content/10.1...

Generative design of novel bacteriophages with genome language models www.biorxiv.org/content/10.1...
Leveraging Evo 1 & Evo 2, they generated whole genome sequences, resulting in 16 viable phages with distinct genomic architectures.

Leveraging Evo 1 & Evo 2, they generated whole genome sequences, resulting in 16 viable phages with distinct genomic architectures.
Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
A new National Academies report examines how AI-driven biological tools could impact #biosecurity, from potential misuse to their role in risk mitigation.
Read: buff.ly/2unawE4

A new National Academies report examines how AI-driven biological tools could impact #biosecurity, from potential misuse to their role in risk mitigation.
Read: buff.ly/2unawE4
"Genome modeling and design across all domains of life with Evo 2" youtu.be/Rarn97Wpl1A
With the author Garyk Brixi!

"Genome modeling and design across all domains of life with Evo 2" youtu.be/Rarn97Wpl1A
With the author Garyk Brixi!
@garykbrixi.bsky.social @matthewdurrant.com @michaelpoli.bsky.social @genophoria.bsky.social @pdhsu.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...

@garykbrixi.bsky.social @matthewdurrant.com @michaelpoli.bsky.social @genophoria.bsky.social @pdhsu.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
@richardshuai.bsky.social Talal Widatalla @possuhuanglab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
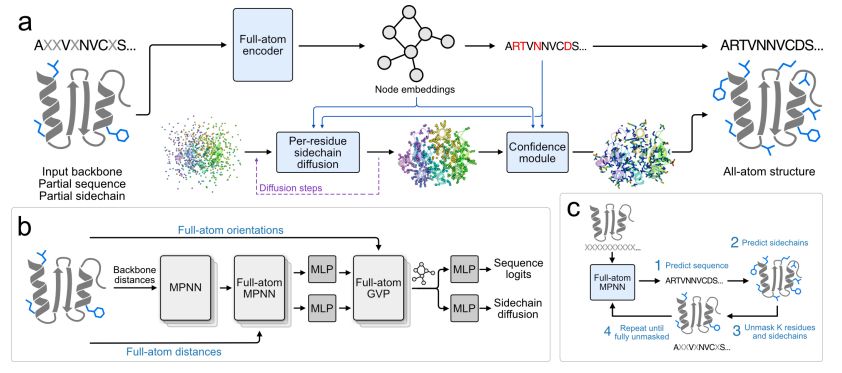
@richardshuai.bsky.social Talal Widatalla @possuhuanglab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
The model achieves an unprecedented breadth in capabilities, enabling prediction and design tasks from molecular to genome scale and across all three domains of life.

The model achieves an unprecedented breadth in capabilities, enabling prediction and design tasks from molecular to genome scale and across all three domains of life.

nytimes.com/2024/12/30/t...
congrats!
@arcinstitute.org @brianhie.bsky.social @patrickhsuu.bsky.social @jameszou.bsky.social and the others for well deserved recognition

nytimes.com/2024/12/30/t...
congrats!
@arcinstitute.org @brianhie.bsky.social @patrickhsuu.bsky.social @jameszou.bsky.social and the others for well deserved recognition
@brianhie.bsky.social

@brianhie.bsky.social

- toxin/antitoxin pairs
- anti-CRISPR proteins
+ a database of 120B synthetic base pairs
www.biorxiv.org/content/10.1...
@brianhie.bsky.social
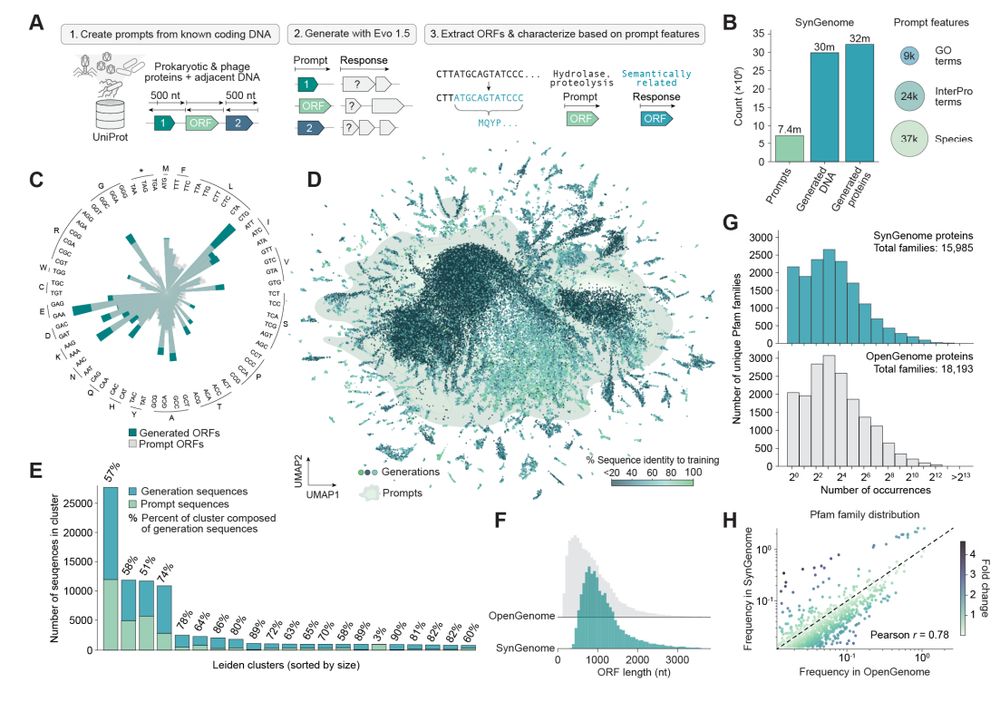
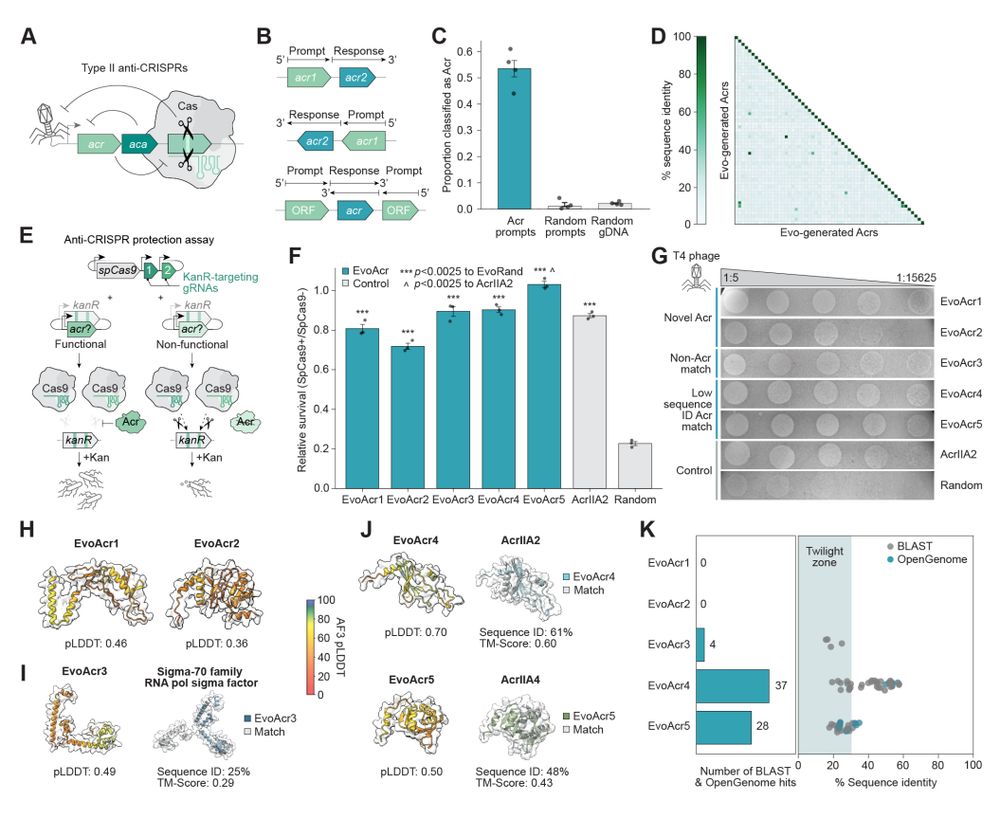
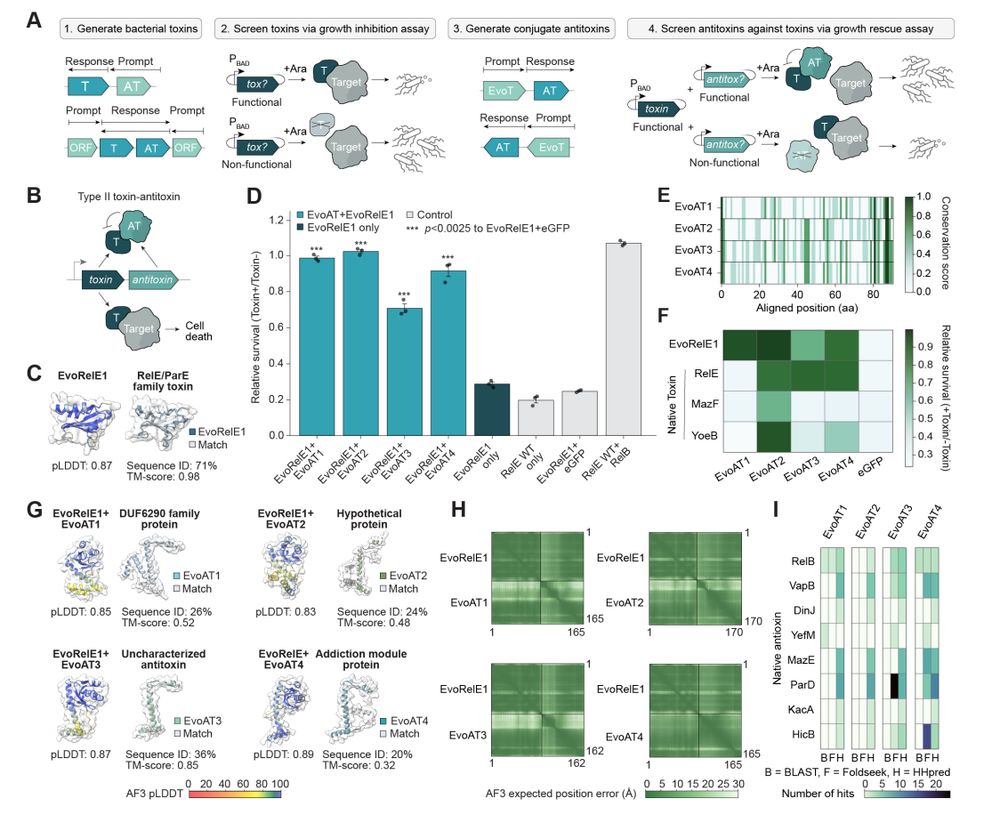
- toxin/antitoxin pairs
- anti-CRISPR proteins
+ a database of 120B synthetic base pairs
www.biorxiv.org/content/10.1...
@brianhie.bsky.social

