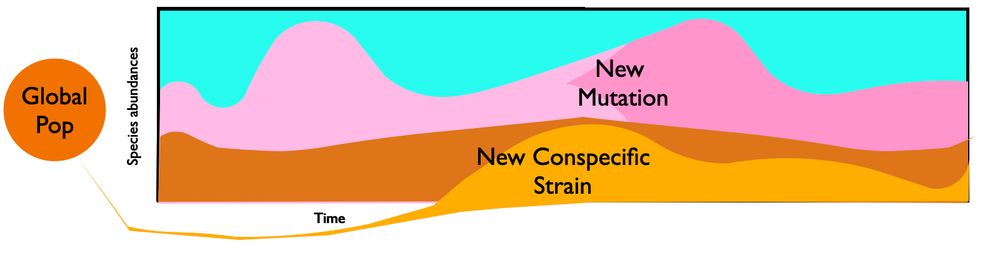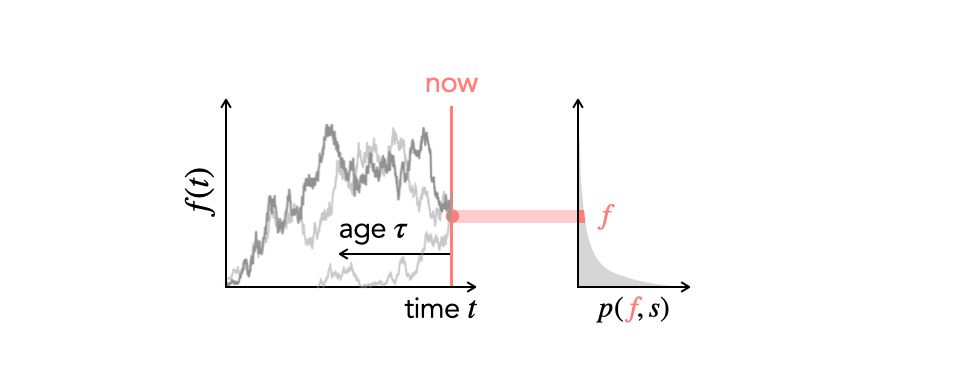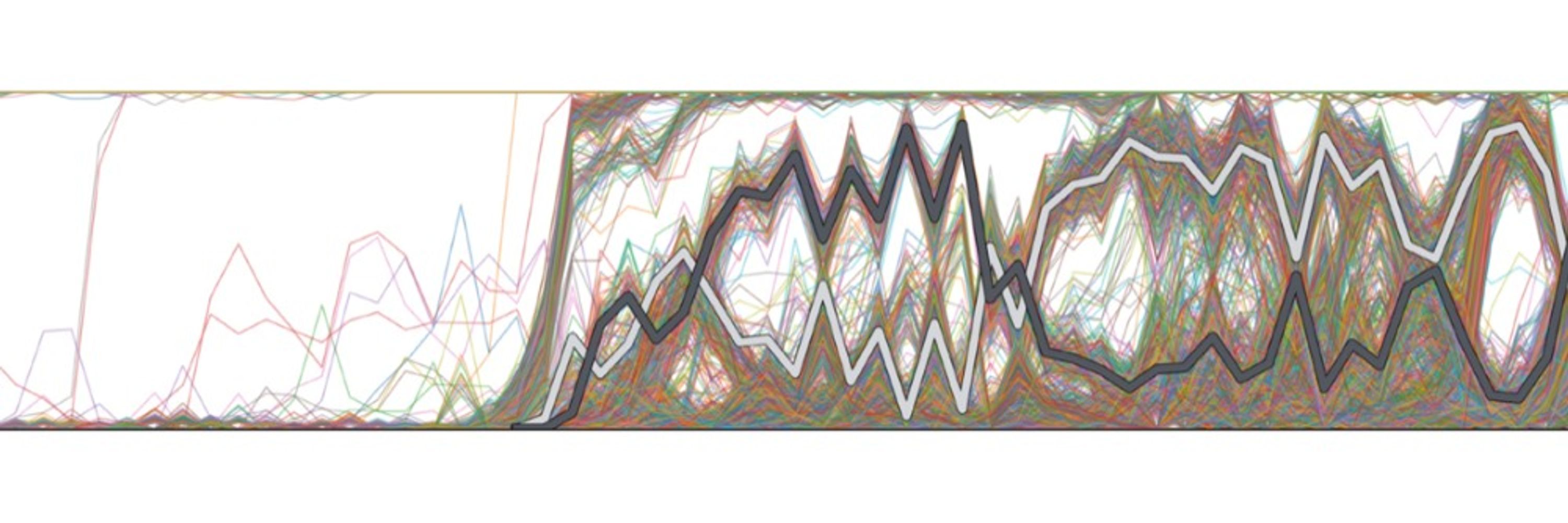
www.biorxiv.org/content/10.1...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...

apply.interfolio.com/177622
Suggested deadline: 12/15/2025.
@columbiasysbio.bsky.social

apply.interfolio.com/177622
Suggested deadline: 12/15/2025.
@columbiasysbio.bsky.social
A solid machine learning framework & to predict strain-level phage-host interactions across diverse bacterial genera from genome sequences alone. Avery Noonan from the Arkin Lab led this massive effort
www.biorxiv.org/content/10.1...
A solid machine learning framework & to predict strain-level phage-host interactions across diverse bacterial genera from genome sequences alone. Avery Noonan from the Arkin Lab led this massive effort
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
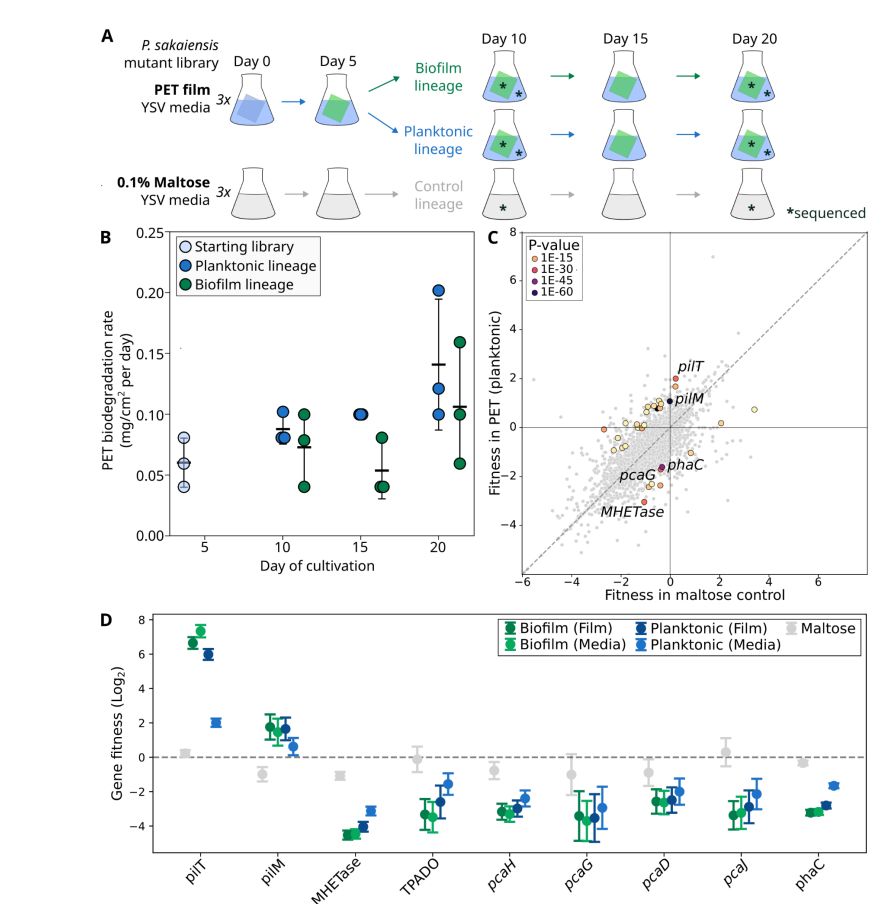
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
www.cell.com/trends/micro...

www.cell.com/trends/micro...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
1) MMG (my dept): fundamental research in med micro
2) Peds ID / I4Kids institute
3) Center for Vaccine Research
🔗 to all 3 w/info: www.linkedin.com/posts/vaughn...

1) MMG (my dept): fundamental research in med micro
2) Peds ID / I4Kids institute
3) Center for Vaccine Research
🔗 to all 3 w/info: www.linkedin.com/posts/vaughn...
If you’re interested in population genetics, fitness landscapes, and viral evolution — get in touch.
faculty-emory.icims.com/jobs/151181/...
If you’re interested in population genetics, fitness landscapes, and viral evolution — get in touch.
faculty-emory.icims.com/jobs/151181/...

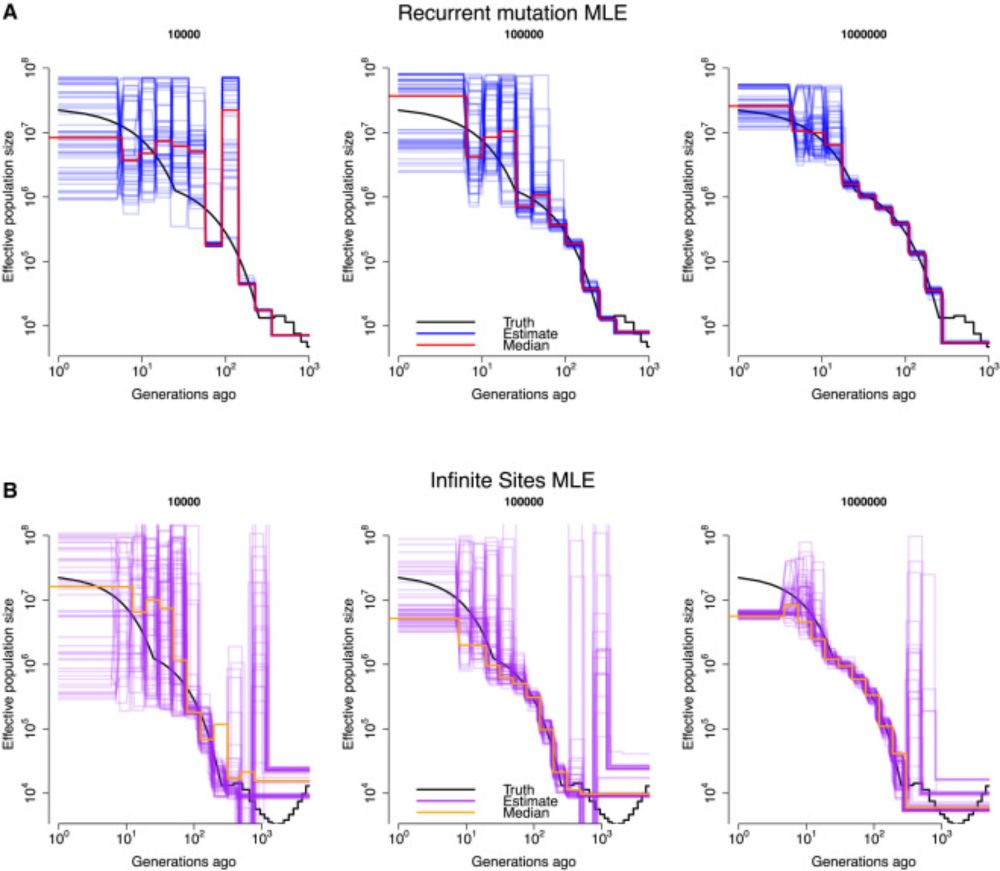
📄Estimation of demography and mutation rates from one million haploid genomes
🧑🤝🧑 @jgschraiber.bsky.social @jeffspence.github.io @docedge.bsky.social
Excellent work by UW Master's student Gechlang Tang in @asm.org #mSystems Journal.
journals.asm.org/doi/10.1128/...
🧵
Excellent work by UW Master's student Gechlang Tang in @asm.org #mSystems Journal.
journals.asm.org/doi/10.1128/...
🧵
recruit.ap.uci.edu/JPF09601

recruit.ap.uci.edu/JPF09601

I am looking for a computational scientist to join my genomics lab at Stanford. They should have an outstanding skillset in ML/statistical methods for genomic applications, postdoc experience and a strong publication record.
#sciencejobs
I am looking for a computational scientist to join my genomics lab at Stanford. They should have an outstanding skillset in ML/statistical methods for genomic applications, postdoc experience and a strong publication record.
#sciencejobs
www.biorxiv.org/content/10.1...