
➡️ https://l.humanite.fr/nqu

➡️ https://l.humanite.fr/nqu
py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)

py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)
Trailer for the documentary:
www.instagram.com/reel/DQumf36...
Trailer for the documentary:
www.instagram.com/reel/DQumf36...

Please contact Nathalie Lagarde for more details ([email protected])

Please contact Nathalie Lagarde for more details ([email protected])
Not unexpected, given the disastrous policy changes in the US, but nonetheless horrible news.
Exclusive: Famed protein structure competition nears end as NIH grant money runs out | Science | AAAS
Not unexpected, given the disastrous policy changes in the US, but nonetheless horrible news.
Exclusive: Famed protein structure competition nears end as NIH grant money runs out | Science | AAAS
www.understandingai.org/p/i-got-fool...

www.understandingai.org/p/i-got-fool...
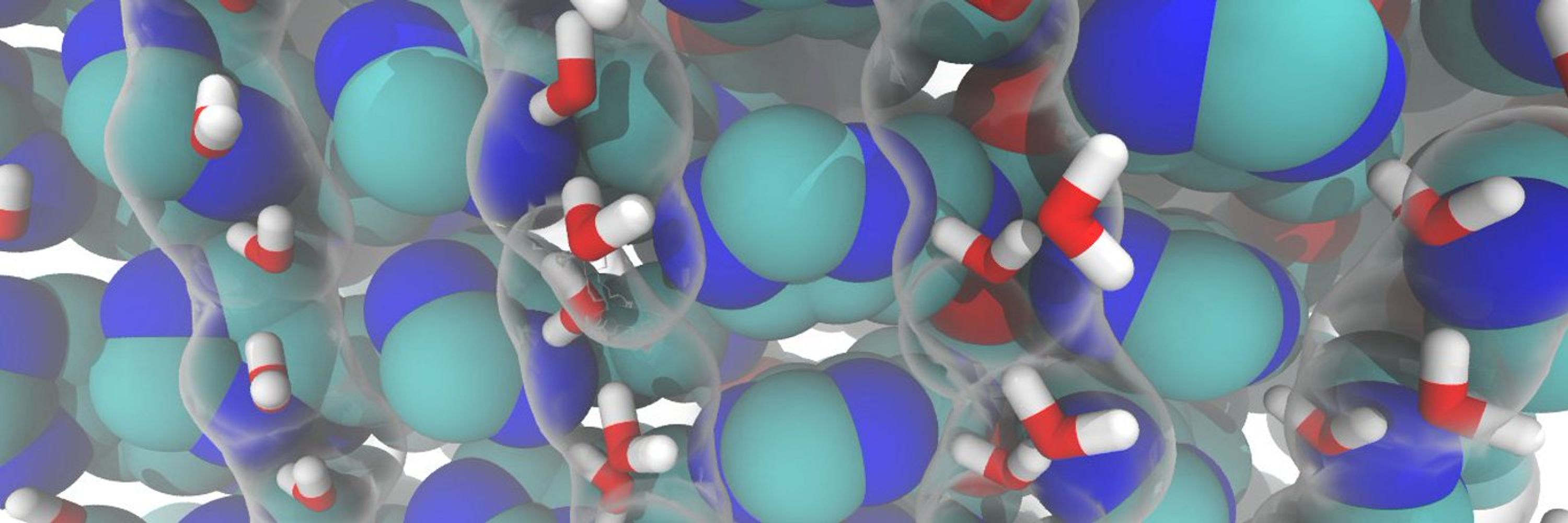
Accelerates biomolecule simulations by scaling solute interactions, enhancing structural sampling at fewer temperatures.




It is now possible to add post-translational modifications and to run ligand screening with AlphaFold3!

It is now possible to add post-translational modifications and to run ligand screening with AlphaFold3!
Ah bon ? Et pourquoi pas le reste ?
Pourquoi pas pour détournement de fonds publics ? »
— Marine Le Pen, le 5 avril 2013.
#PassionArchives
Ah bon ? Et pourquoi pas le reste ?
Pourquoi pas pour détournement de fonds publics ? »
— Marine Le Pen, le 5 avril 2013.
#PassionArchives
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.nature.com/artic...
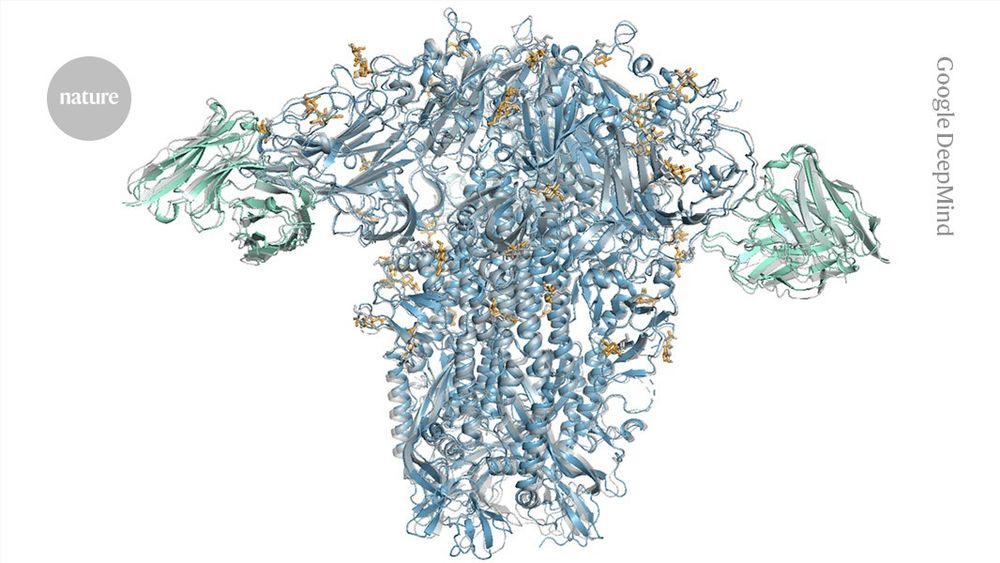
www.nature.com/artic...
🔬✨ This open-source tool streamlines the analysis of protein structures, offering functions to compare, visualize, and evaluate model quality.
🔬✨ This open-source tool streamlines the analysis of protein structures, offering functions to compare, visualize, and evaluate model quality.



www.ks.uiuc.edu/Research/vmd...

www.ks.uiuc.edu/Research/vmd...


www.lemonde.fr/idees/articl...

www.lemonde.fr/idees/articl...
www.biorxiv.org/content/10.1...
🧵👇 (1/n)

www.biorxiv.org/content/10.1...
🧵👇 (1/n)
1/3 des fermetures de classe concentrées sur 3 arrondissements populaires.
20minutes
www.20minutes.fr/paris/413747...

1/3 des fermetures de classe concentrées sur 3 arrondissements populaires.


