@imbavienna.bsky.social solved a decade-old puzzle, uncovering how the information molecule mRNA travels from the cell’s nucleus to its periphery. More: bit.ly/4nHcvys

@imbavienna.bsky.social solved a decade-old puzzle, uncovering how the information molecule mRNA travels from the cell’s nucleus to its periphery. More: bit.ly/4nHcvys
what started as a project on how cells export piRNA precursors, ended up as a tour de force in mRNA export. truly wonderful collaboration with @plaschkalab.bsky.social at the @viennabiocenter.bsky.social
@imbavienna.bsky.social solved a decade-old puzzle, uncovering how the information molecule mRNA travels from the cell’s nucleus to its periphery. More: bit.ly/4nHcvys

what started as a project on how cells export piRNA precursors, ended up as a tour de force in mRNA export. truly wonderful collaboration with @plaschkalab.bsky.social at the @viennabiocenter.bsky.social
We asked how can protein complexes diversify without compromising their function and explored this question using the plant #exocyst complex.
www.nature.com/articles/s41...

We asked how can protein complexes diversify without compromising their function and explored this question using the plant #exocyst complex.
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
#teamtomo

www.biorxiv.org/content/10.1...
#teamtomo
We will explore new mechanisms in eukaryotic gene expression, leveraging ‘evolutionary play’ to uncover how regulation, repurposing, and hijacking shape RNA biology.
PhD positions available!
We will explore new mechanisms in eukaryotic gene expression, leveraging ‘evolutionary play’ to uncover how regulation, repurposing, and hijacking shape RNA biology.
PhD positions available!
#cryoEM

#cryoEM


More info: www.imb.de/students-pos...
More info: www.imb.de/students-pos...
www.cell.com/cell/fulltex...

www.cell.com/cell/fulltex...
www.nature.com/articles/s41...
www.nature.com/articles/s41...


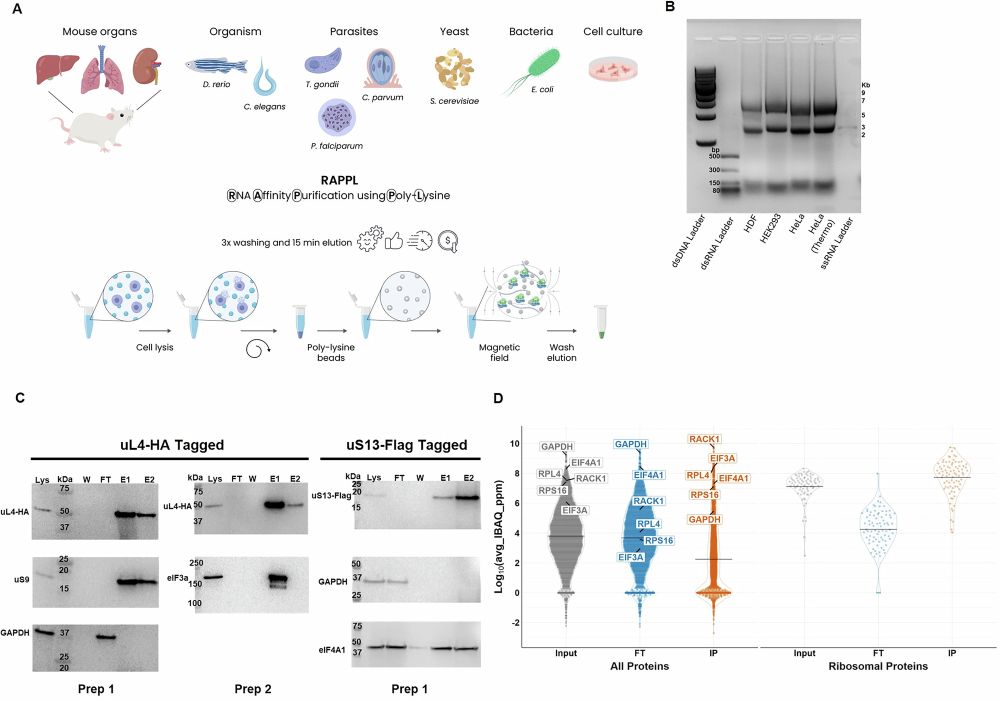
A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo

A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo
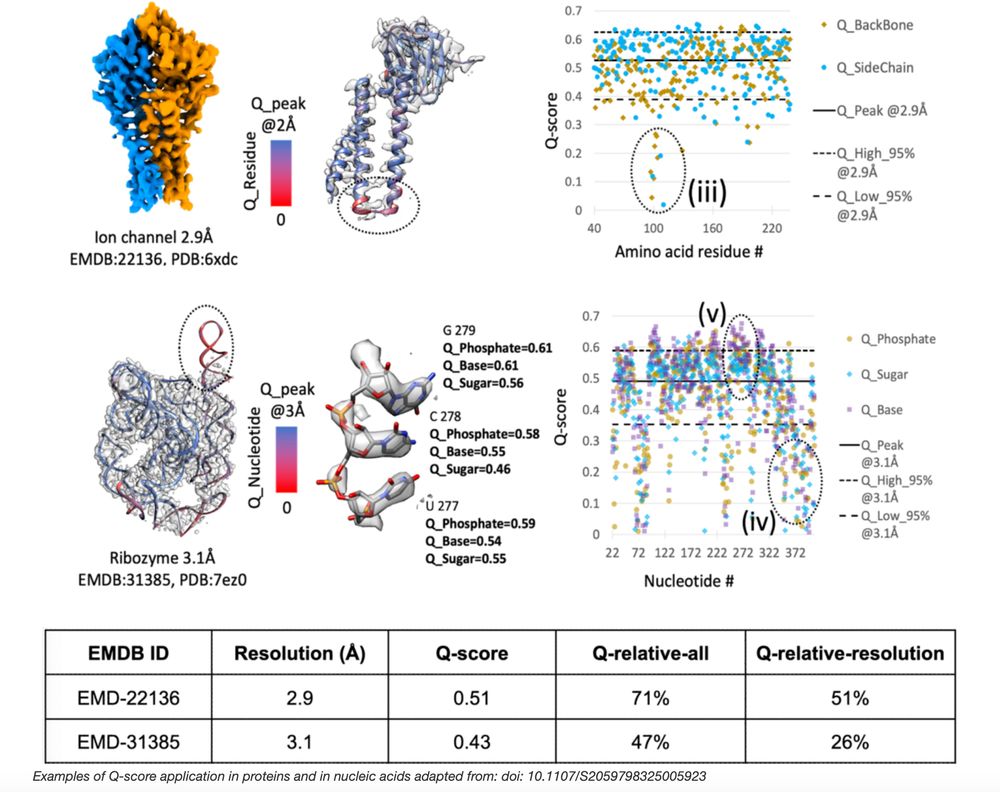
rdcu.be/ezlhg
rdcu.be/ezlhg
biorxiv.org/content/10.110
biorxiv.org/content/10.110
the Liu group we report the development of a laboratory-evolved CRISPR-associated transposase (evoCAST) that supports therapeutically relevant levels of RNA-programmable gene insertion in human cells. drive.google.com/file/d/1I-Ub...
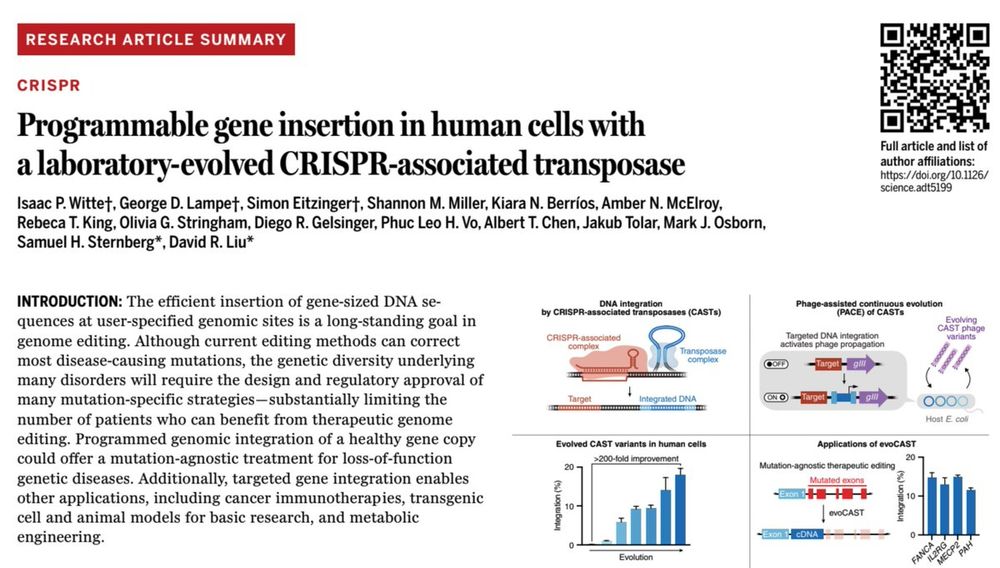
the Liu group we report the development of a laboratory-evolved CRISPR-associated transposase (evoCAST) that supports therapeutically relevant levels of RNA-programmable gene insertion in human cells. drive.google.com/file/d/1I-Ub...

I am especially honoured that my poster was awarded with one of the two poster prizes 😇


I am especially honoured that my poster was awarded with one of the two poster prizes 😇


⏳ Deadline: April 15 🚨, apply now: training.vbc.ac.at/phd-program/
📽️Watch David's 'Scientist Snapshot': youtube.com/watch?v=DXTO...
@haselbachlab.bsky.social

It was a pleasure seeing this project unfold 🚀
Let me walk you through it 🧵 1/9

It was a pleasure seeing this project unfold 🚀