Julian Streit
@julianstreit.bsky.social
64 followers
58 following
6 posts
Postdoctoral researcher in computational structural biology at the University of Copenhagen with Kresten Lindorff-Larsen
Posts
Media
Videos
Starter Packs
Reposted by Julian Streit
Reposted by Julian Streit
Chris Waudby
@chriswaudby.bsky.social
· Sep 24
Kinetic NMR screening: rapidly quantifying fast ligand dissociation in fragment mixtures using ¹⁹F relaxation dispersion
Ligand-observed NMR is a key tool for detecting weak protein-ligand interactions, but translating binding signals into meaningful affinity measurements remains difficult, limiting the ability to rank ...
doi.org
Reposted by Julian Streit
Giulio Tesei
@giuliotesei.bsky.social
· Sep 18
New review on computational design of intrinsically disordered proteins 🖥️🍝 by @giuliotesei.bsky.social @fpesce.bsky.social & 👴
doi.org/10.48550/arX...
doi.org/10.48550/arX...

Reposted by Julian Streit
Julian Streit
@julianstreit.bsky.social
· Jun 28

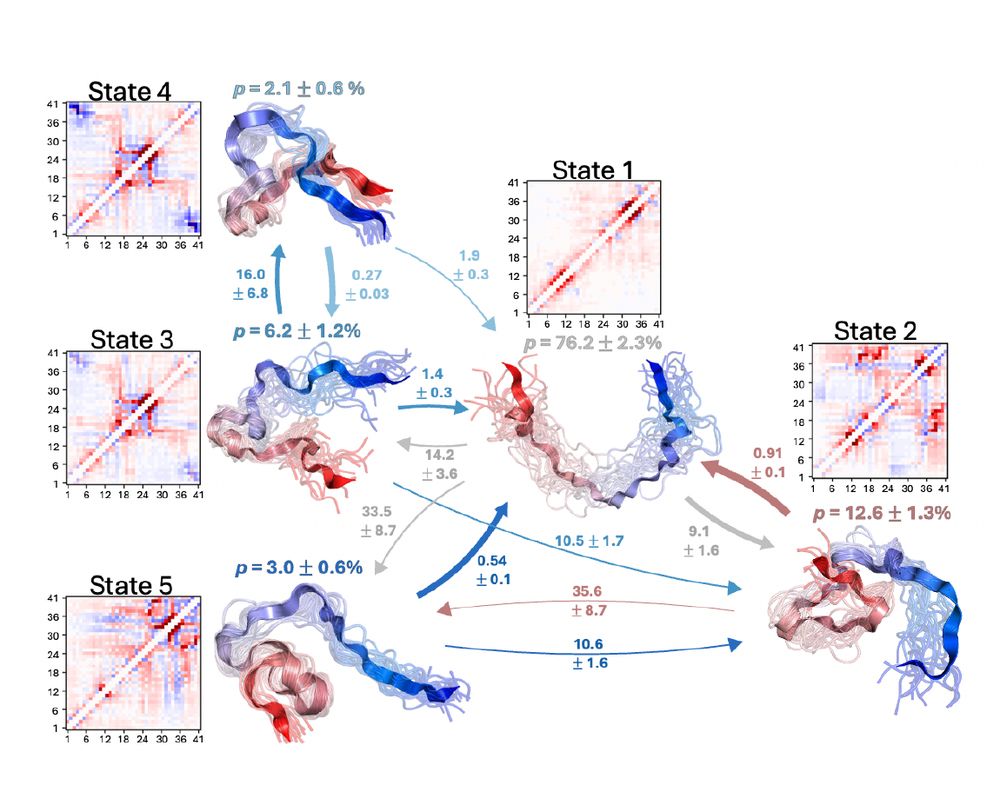
Protein Dynamics at Different Timescales Unlock Access to Hidden Post-Translational Modification Sites
Post-translational modifications (PTMs) alter the proteome in response to intra- and extracellular signals, providing fundamental information processing in development, homeostasis and disease. Here, ...
biorxiv.org
Julian Streit
@julianstreit.bsky.social
· Jun 25
In press!! NMR & MD simulations reveal how non-native contacts stabilise a key transient intermediate in the lysozyme amyloid cascade. Well done Minkoo and all!! @phar.cam.ac.uk For CMD ❤️ advanced.onlinelibrary.wiley.com/doi/10.1002/...

Amyloid Forming Human Lysozyme Intermediates are Stabilized by Non‐Native Amide‐π Interactions
Mutational variants of human lysozyme cause fatal systemic amyloidosis by depositing kilograms of protein in the viscera of patients. Central to this process is a partially unfolded protein intermedi....
advanced.onlinelibrary.wiley.com
Reposted by Julian Streit
Janet Kumita
@jrkumita.bsky.social
· Jun 25

Amyloid Forming Human Lysozyme Intermediates are Stabilized by Non‐Native Amide‐π Interactions
Mutational variants of human lysozyme cause fatal systemic amyloidosis by depositing kilograms of protein in the viscera of patients. Central to this process is a partially unfolded protein intermedi....
advanced.onlinelibrary.wiley.com
Reposted by Julian Streit
Reposted by Julian Streit
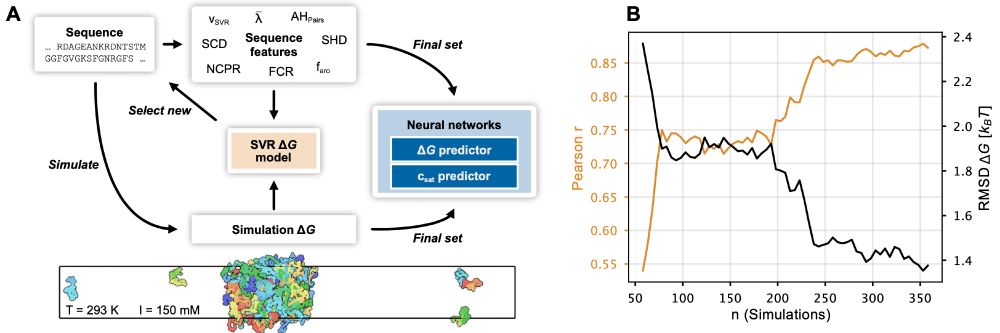
In collaboration with the @lindorfflarsen.bsky.social group we release our map of degrons in >5,000 human cytosolic proteins with >99% coverage. A machine learning model trained on the data identifies missense variants forming degrons in exposed & disordered regions. Work led by @vvouts.bsky.social.

A complete map of human cytosolic degrons and their relevance for disease
Degrons are short protein segments that target proteins for degradation via the ubiquitin-proteasome system and thus ensure timely removal of signaling proteins and clearance of misfolded proteins fro...
www.biorxiv.org
Reposted by Julian Streit
Sammy Chan
@sammyhschan.bsky.social
· May 8
Reposted by Julian Streit
Reposted by Julian Streit
Julian Streit
@julianstreit.bsky.social
· Apr 11
Reposted by Julian Streit
Reposted by Julian Streit
Sammy Chan
@sammyhschan.bsky.social
· Apr 10
The ribosome directs nascent chains through two folding-dependent pathways https://www.biorxiv.org/content/10.1101/2025.04.08.647855v1
Reposted by Julian Streit
Reposted by Julian Streit
Reposted by Julian Streit
Reposted by Julian Streit
Reposted by Julian Streit
Sammy Chan
@sammyhschan.bsky.social
· Feb 15

Long-range electrostatic forces govern how proteins fold on the ribosome
Protein biosynthesis and folding are tightly intertwined processes regulated by the ribosome and auxiliary factors. Nascent proteins can begin to fold on their parent ribosome but formation of the nat...
doi.org
Julian Streit
@julianstreit.bsky.social
· Feb 14
Julian Streit
@julianstreit.bsky.social
· Feb 14

Long-range electrostatic forces govern how proteins fold on the ribosome
Protein biosynthesis and folding are tightly intertwined processes regulated by the ribosome and auxiliary factors. Nascent proteins can begin to fold on their parent ribosome but formation of the nat...
www.biorxiv.org