
Berkeley, CA
CinderBio.com

Come see posters using CinderBio enzymes on the schedule and topics below.
Steven Yannone our Founder and CEO, will be at the conference. Reach out to [email protected] for more information.
#proteomics #TeamMassSpec
#asms2025 #proteomics
@hecklab.bsky.social @tshamorkina.bsky.social @tkadava.bsky.social @joostsnijder.bsky.social
www.biorxiv.org/content/10.1...

#asms2025 #proteomics
@hecklab.bsky.social @tshamorkina.bsky.social @tkadava.bsky.social @joostsnijder.bsky.social
It was a true team effort @lperezpaneda.bsky.social @tkadava.bsky.social @joostsnijder.bsky.social and @hecklab.bsky.social
---
#proteomics #prot-preprint
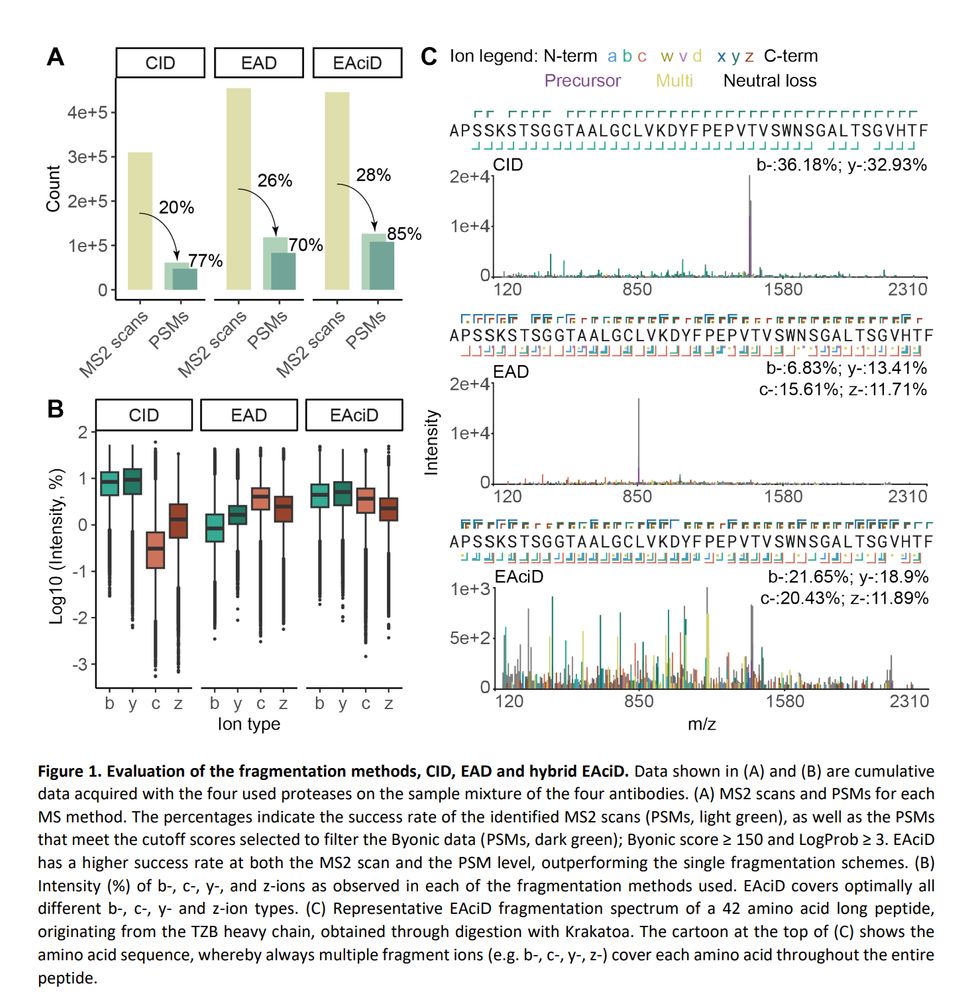
It was a true team effort @lperezpaneda.bsky.social @tkadava.bsky.social @joostsnijder.bsky.social and @hecklab.bsky.social
Come see posters using CinderBio enzymes on the schedule and topics below.
Steven Yannone our Founder and CEO, will be at the conference. Reach out to [email protected] for more information.
#proteomics #TeamMassSpec

Come see posters using CinderBio enzymes on the schedule and topics below.
Steven Yannone our Founder and CEO, will be at the conference. Reach out to [email protected] for more information.
#proteomics #TeamMassSpec
---
#proteomics #prot-paper

---
#proteomics #prot-paper
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...

A super collaboration trying to solve a bottleneck in diagnostic medicine.
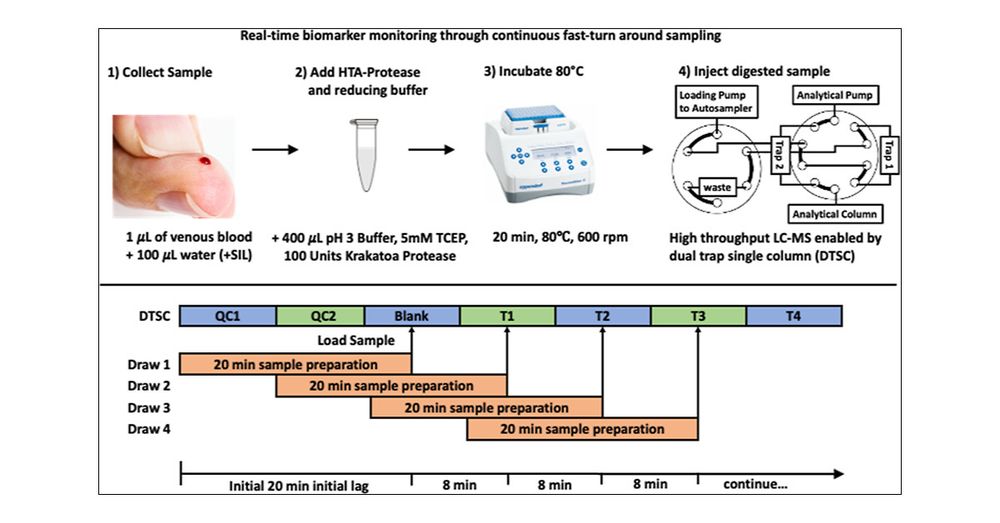
A super collaboration trying to solve a bottleneck in diagnostic medicine.

cinderbio.com
#Proteomics #TeamMassSpec

cinderbio.com
#Proteomics #TeamMassSpec
#proteomics

#proteomics


