BPNet/ChromBPNet are powerful models for understanding regulatory genomics from @anshulkundaje.bsky.social's group, and now it's way easier to go from raw data to trained models and analysis + results in PyTorch
Try it out with `pip install bpnet-lite`
BPNet/ChromBPNet are powerful models for understanding regulatory genomics from @anshulkundaje.bsky.social's group, and now it's way easier to go from raw data to trained models and analysis + results in PyTorch
Try it out with `pip install bpnet-lite`

Section 8, listing properties unique to neural networks, is quite interesting.

Section 8, listing properties unique to neural networks, is quite interesting.

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
A new, comprehensive study of 263 species documents higher cancer prevalence with increasing body mass. Some large animals (e.g. elephants) have some built-in genetic adaptations
www.pnas.org/doi/10.1073/... @pnas.org
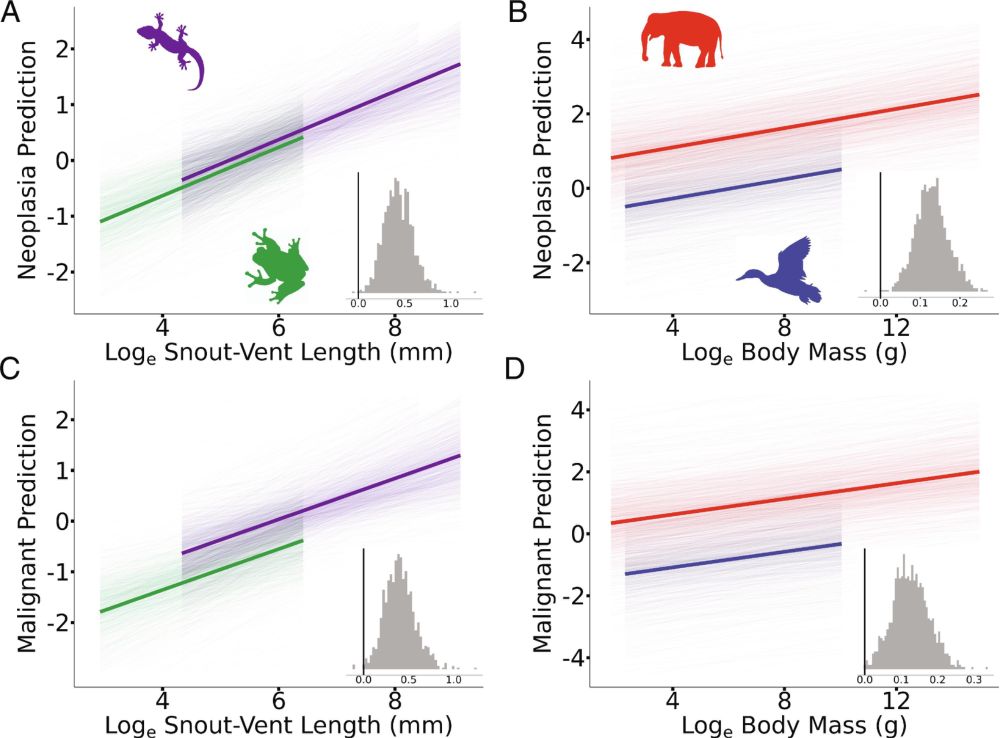
A new, comprehensive study of 263 species documents higher cancer prevalence with increasing body mass. Some large animals (e.g. elephants) have some built-in genetic adaptations
www.pnas.org/doi/10.1073/... @pnas.org
Today, @arcinstitute.org in collaboration with Nvidia releases Evo 2—a fully open source biological foundation model trained on genomes spanning the entire tree of life.
academic.oup.com/nar/article/...

academic.oup.com/nar/article/...
Lots of interesting new TE (& genes) biology
Data fully browsable💻 👉 embryo.helmholtz-munich.de/shiny_embryo/
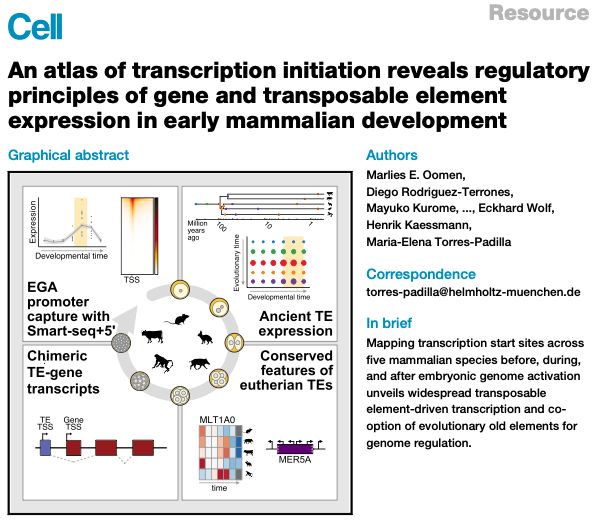
Lots of interesting new TE (& genes) biology
Data fully browsable💻 👉 embryo.helmholtz-munich.de/shiny_embryo/
hocomoco.autosome.org

hocomoco.autosome.org
▶️40-fold flanking sequence effect on TF binding
▶️Motif recognition beats kinetics
▶️Minutes-long residence times
▶️Models of TF-nucleosome competiton






www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
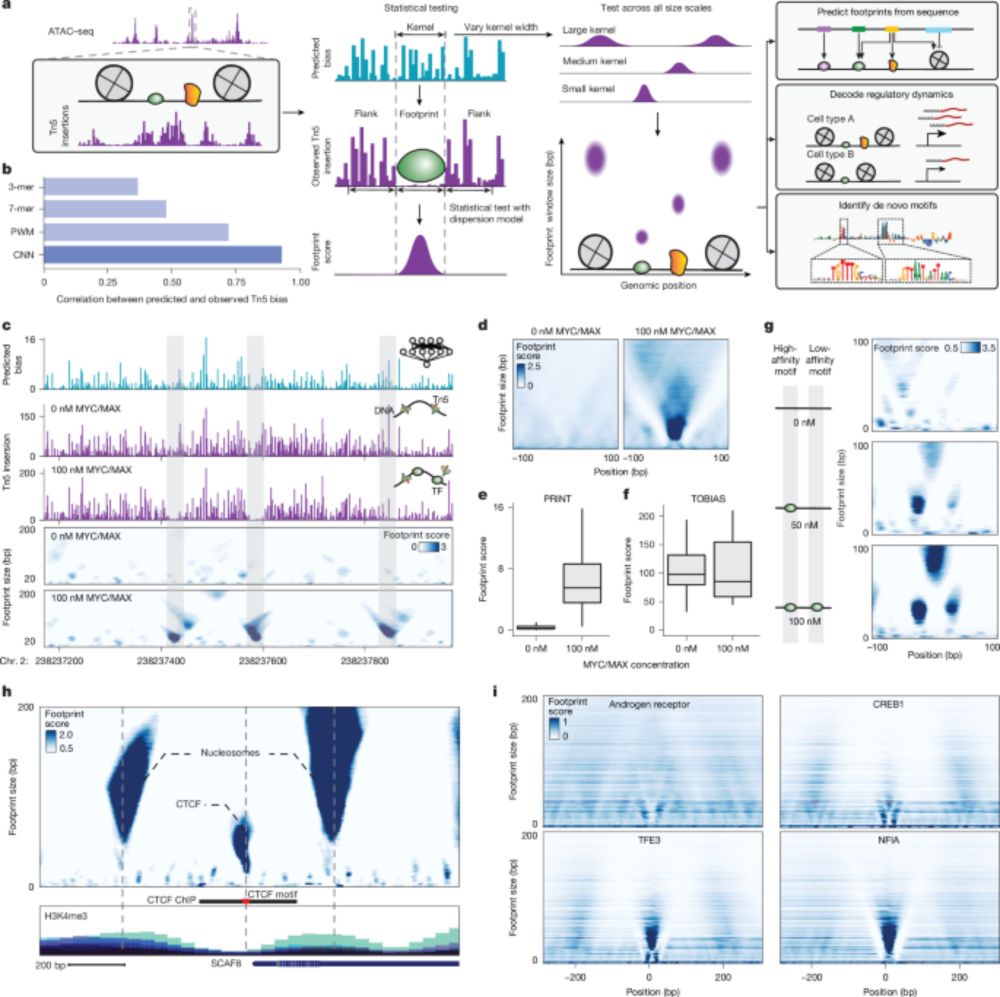
www.nature.com/articles/s41...


scCustomize sets some better defaults that help with lots of the issues. :)
samuel-marsh.github.io/scCustomize/...
scCustomize sets some better defaults that help with lots of the issues. :)
samuel-marsh.github.io/scCustomize/...