
Researcher @ University of Oslo
#MicroSky #IDSky #Protistsonsky 🧬💻
www.microbiologyresearch.org/content/micr...
#MicroSky #IDSky #Protistsonsky 🧬💻
www.microbiologyresearch.org/content/micr...
We found that hospital patients were frequently colonised with P. aeruginosa and that the same clone was shared between the gut and the lung.
The phylogenies indicate that the clones moved from lung->gut
www.nature.com/articles/s41...
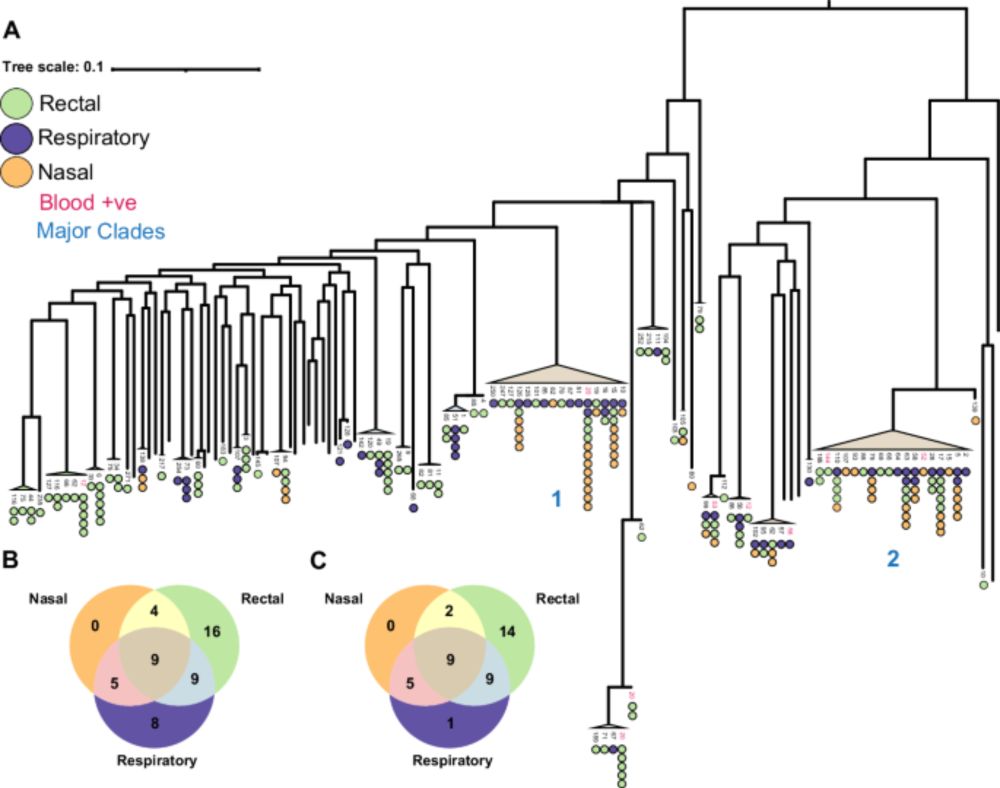
We found that hospital patients were frequently colonised with P. aeruginosa and that the same clone was shared between the gut and the lung.
The phylogenies indicate that the clones moved from lung->gut
www.nature.com/articles/s41...
Anyone who’s tried deleting prophages in the lab by HR knows the difficulties of the task. Here we have an example of how HR-mediated natural transformation might hit the same hurdle in a more native context.
academic.oup.com/mbe/article/...

Anyone who’s tried deleting prophages in the lab by HR knows the difficulties of the task. Here we have an example of how HR-mediated natural transformation might hit the same hurdle in a more native context.
academic.oup.com/mbe/article/...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
journals.plos.org/plosbiology/...

journals.plos.org/plosbiology/...
We're excited for users who might train new models, find phenotype/genotype mismatches, or any other use
EMBL-EBI’s new AMR portal brings together laboratory resistance data and bacterial genomes in one open platform.
#WAAW2025 #ActOnAMR
www.ebi.ac.uk/about/news/t...
🧬💻

We're excited for users who might train new models, find phenotype/genotype mismatches, or any other use

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
4 yrs on, even Nextstrain has lost access. GISAID has rotted from its core.

4 yrs on, even Nextstrain has lost access. GISAID has rotted from its core.
Read the full opinion piece in @cp-neuron.bsky.social: spkl.io/63322AbxpA
@wiringthebrain.bsky.social, @statsepi.bsky.social, & @deevybee.bsky.social

Read the full opinion piece in @cp-neuron.bsky.social: spkl.io/63322AbxpA
@wiringthebrain.bsky.social, @statsepi.bsky.social, & @deevybee.bsky.social
I'm looking for studies where both Illumina and ONT sequencing were performed on the same samples from soil, human, ruminent, and other sample types for comparison. Bonus if those studies include PacBio data.
Please help and share!
I'm looking for studies where both Illumina and ONT sequencing were performed on the same samples from soil, human, ruminent, and other sample types for comparison. Bonus if those studies include PacBio data.
Please help and share!
Excellent ‘genomics in context’ call from @wellcometrust.bsky.social
wellcome.org/research-fun...

Excellent ‘genomics in context’ call from @wellcometrust.bsky.social
wellcome.org/research-fun...
The ONE CONSTANT in the wake of the serial crisis, #PlanS and #OpenAccess reform has been publish profit margins.
2/n

The ONE CONSTANT in the wake of the serial crisis, #PlanS and #OpenAccess reform has been publish profit margins.
2/n
a 🧵 1/n
Drain: arxiv.org/abs/2511.04820
Strain: direct.mit.edu/qss/article/...
Oligopoly: direct.mit.edu/qss/article/...




a 🧵 1/n
Drain: arxiv.org/abs/2511.04820
Strain: direct.mit.edu/qss/article/...
Oligopoly: direct.mit.edu/qss/article/...
Join a world-class biomedical research institute at the heart of the Vienna BioCenter, where curiosity drives discovery.
Lead your own lab, pursue bold ideas, and shape the future of science at the IMP: www.imp.ac.at/career/open-...

Join a world-class biomedical research institute at the heart of the Vienna BioCenter, where curiosity drives discovery.
Lead your own lab, pursue bold ideas, and shape the future of science at the IMP: www.imp.ac.at/career/open-...
Cool @natmicrobiol.nature.com publication by @erikbakkeren.bsky.social
@vit-pi.bsky.social @meganleeny.bsky.social @microscape.bsky.social & Kevin Foster
www.nature.com/articles/s41...

Cool @natmicrobiol.nature.com publication by @erikbakkeren.bsky.social
@vit-pi.bsky.social @meganleeny.bsky.social @microscape.bsky.social & Kevin Foster
www.nature.com/articles/s41...
For the first time, scientists can predict the rate at which gut bacteria pass between people — a major step towards tackling antibiotic-resistant infections.
Read here ⤵️
sanger.ac.uk/news_item/ad...

For the first time, scientists can predict the rate at which gut bacteria pass between people — a major step towards tackling antibiotic-resistant infections.
Read here ⤵️
sanger.ac.uk/news_item/ad...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Comes with 5 years protected research time, a PhD studentship, & 60k start-up. I had one of these before getting my FLF, happy to chat to anyone interested!
www.qub.ac.uk/Research/fel...
Comes with 5 years protected research time, a PhD studentship, & 60k start-up. I had one of these before getting my FLF, happy to chat to anyone interested!
www.qub.ac.uk/Research/fel...
It said a restriction map was "extremely unlikely to have arisen by random evolution.”
I took its claims seriously.
The "synthetic fingerprint" hypothesis collapses. My new preprint explains why.
arxiv.org/abs/2510.23833

It said a restriction map was "extremely unlikely to have arisen by random evolution.”
I took its claims seriously.
The "synthetic fingerprint" hypothesis collapses. My new preprint explains why.
arxiv.org/abs/2510.23833


