My interests are epigenomics, chromatin biology, gene regulation, machine learning and causal inference
https://ntanmayee.github.io/
Histone modifications are measured with ChIP-Seq, but the chromatin landscape introduces biases. We introduce DecoDen to simultaneously learn shared chromatin landscapes while de-biasing individual measurement tracks.
Check it out: github.com/ntanmayee/de...
🧬 🖥️
Some results here - arxiv.org/abs/2110.06257

Some results here - arxiv.org/abs/2110.06257

@stuartmac44.bsky.social, @foreveremain.bsky.social, @mitsenkov.bsky.social, Vanessza Fentor, Douglas Freeburn,
@tanmayee.bsky.social and @rosiegallagher.bsky.social!

@stuartmac44.bsky.social, @foreveremain.bsky.social, @mitsenkov.bsky.social, Vanessza Fentor, Douglas Freeburn,
@tanmayee.bsky.social and @rosiegallagher.bsky.social!
arxiv.org/abs/2110.06257

arxiv.org/abs/2110.06257

Histone modifications are measured with ChIP-Seq, but the chromatin landscape introduces biases. We introduce DecoDen to simultaneously learn shared chromatin landscapes while de-biasing individual measurement tracks.
Check it out: github.com/ntanmayee/de...
🧬 🖥️
Histone modifications are measured with ChIP-Seq, but the chromatin landscape introduces biases. We introduce DecoDen to simultaneously learn shared chromatin landscapes while de-biasing individual measurement tracks.
Check it out: github.com/ntanmayee/de...
🧬 🖥️
Learns shared chromatin structure, correcting histone modification biases across epigenomes.




Learns shared chromatin structure, correcting histone modification biases across epigenomes.
www.huber.embl.de/group/posts/...
www.huber.embl.de/group/posts/...
I've rarely read something so anti-scientific anywhere short of the National Review.
www.nature.com/articles/d41...

I've rarely read something so anti-scientific anywhere short of the National Review.
www.nature.com/articles/d41...
[PhD Position in Computational Evolutionary Transcriptomics]
If you are interested in doing a PhD in gorgeous Scotland on 'Why embryo development goes wrong sometimes?', please consider applying and join our wonderful team in Dundee!
www.dundee.ac.uk/phds/opportu...

[PhD Position in Computational Evolutionary Transcriptomics]
If you are interested in doing a PhD in gorgeous Scotland on 'Why embryo development goes wrong sometimes?', please consider applying and join our wonderful team in Dundee!
www.dundee.ac.uk/phds/opportu...
hocomoco.autosome.org

hocomoco.autosome.org
www.science.org/content/blog...

www.science.org/content/blog...
At best, LLMs gesticulate toward the shoulders of giants."
Bender, West, and I contributed to this pro/con piece in PNAS.

At best, LLMs gesticulate toward the shoulders of giants."
Bender, West, and I contributed to this pro/con piece in PNAS.
Thank you for your kind words about WISR!
Glad to see other organisations like WoAA also mentioned :)

Thank you for your kind words about WISR!
Glad to see other organisations like WoAA also mentioned :)
#DevBio #GeneReg

Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
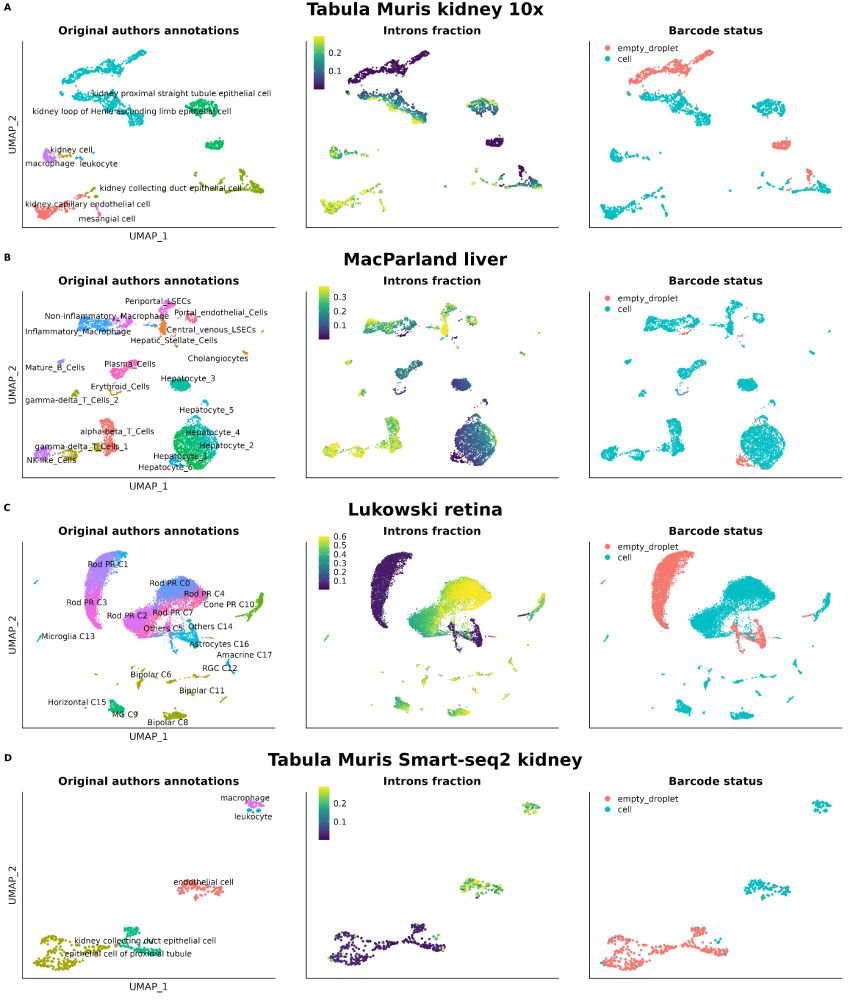
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
www.nature.com/articles/s41...
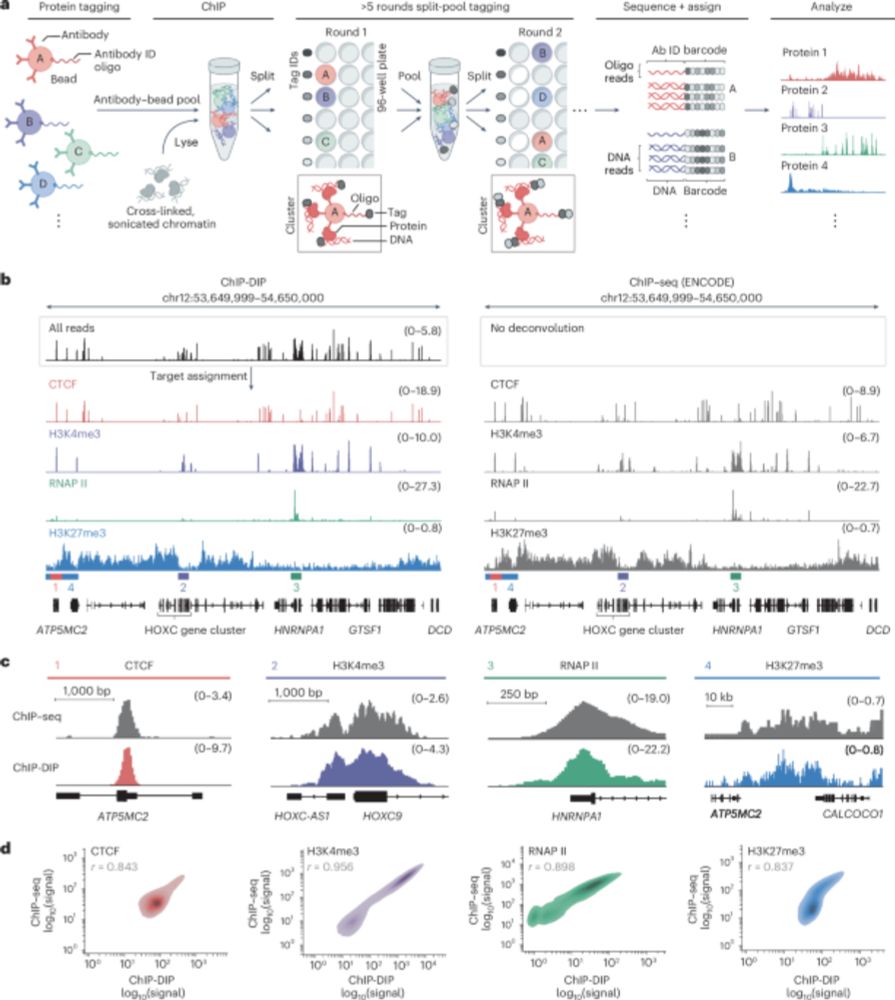
www.nature.com/articles/s41...