
https://nikolai.slavovlab.net

2/n
youtu.be/nIB-WjWzDSc?...
Are protein and phosphorylation levels predictable from genomic and transcriptomic data ?
This study concluded:
"The performance of even the best-performing model was modest, ..."

Are protein and phosphorylation levels predictable from genomic and transcriptomic data ?
This study concluded:
"The performance of even the best-performing model was modest, ..."

I will nominate the first suggestion.
1/2
I will nominate the first suggestion.
1/2
We can profile the diverse pool of modified tRNAs and a wider spectrum of amino acid incorporation into proteins: www.biorxiv.org/content/10.1...
The synergy of these developments will exciting ❗️
1/2

We can profile the diverse pool of modified tRNAs and a wider spectrum of amino acid incorporation into proteins: www.biorxiv.org/content/10.1...
The synergy of these developments will exciting ❗️
1/2
But they are not one thing.
They can have very different generative mechanisms.
Successful analysis starts with understanding their origin and continues with missing aware analysis.
youtu.be/KwprNYv5Pp0?...

But they are not one thing.
They can have very different generative mechanisms.
Successful analysis starts with understanding their origin and continues with missing aware analysis.
youtu.be/KwprNYv5Pp0?...
Keep it in mind the next time you do dimensionality reduction.

Keep it in mind the next time you do dimensionality reduction.
timePlex, PSMtags, and JMod can all support increased throughput for 𝑖𝑛 𝑣𝑖𝑣𝑜 metabolic pulse experiments, and thus make dynamic protein analysis more scalable.
Quote
Mass spec offers a solution.
⬛️ It enables omics scale quantification of the rates of macromolecule synthesis and degradation in a living organism.
blog.slavovlab.net/2025/11/19/i...

timePlex, PSMtags, and JMod can all support increased throughput for 𝑖𝑛 𝑣𝑖𝑣𝑜 metabolic pulse experiments, and thus make dynamic protein analysis more scalable.
Quote
Mass spec offers a solution.
⬛️ It enables omics scale quantification of the rates of macromolecule synthesis and degradation in a living organism.
blog.slavovlab.net/2025/11/19/i...

Mass spec offers a solution.
⬛️ It enables omics scale quantification of the rates of macromolecule synthesis and degradation in a living organism.
blog.slavovlab.net/2025/11/19/i...
It was rejected.
The rejection so dispirited its author, that he did not resubmit it for years.
Now, it's a cornerstone having shaped a field.
It's been cited 78,352 times.
blog.slavovlab.net/2014/08/15/p...

It was rejected.
The rejection so dispirited its author, that he did not resubmit it for years.
Now, it's a cornerstone having shaped a field.
It's been cited 78,352 times.
blog.slavovlab.net/2014/08/15/p...
⬛️ Their abundance strongly declines in AD patients.
=> This decline parallels a reduction in 20S substrate proteins and leads to abnormal protein accumulation in AD.
www.biorxiv.org/content/10.1...

⬛️ Their abundance strongly declines in AD patients.
=> This decline parallels a reduction in 20S substrate proteins and leads to abnormal protein accumulation in AD.
www.biorxiv.org/content/10.1...
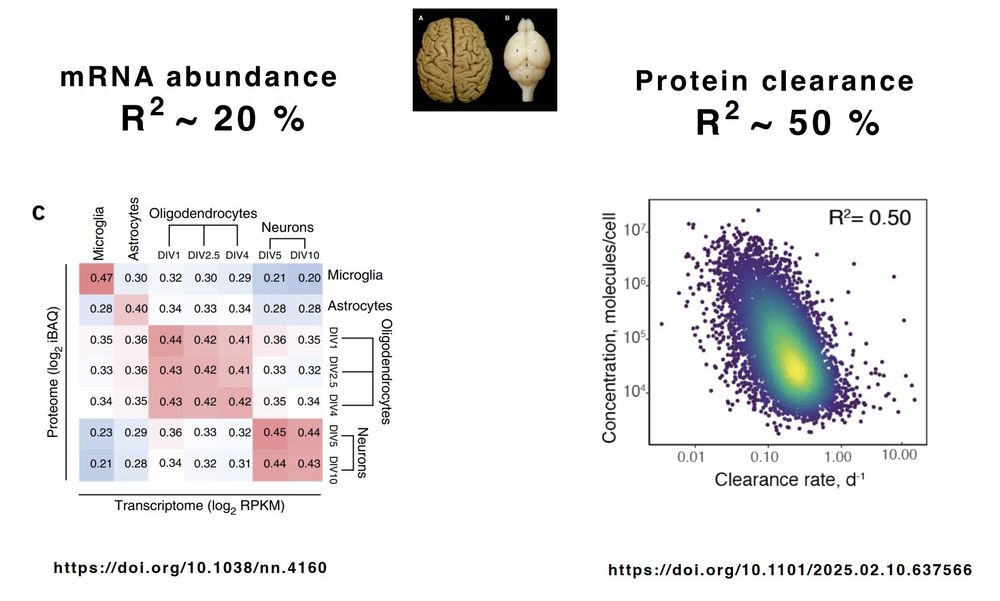
The brain proteome is highly sculpted by protein degradation.
Why ?
This is our model: biorxiv.org/content/10.1...
The brain proteome is highly sculpted by protein degradation.
Why ?
This is our model: biorxiv.org/content/10.1...
In the absence of obvious physical sensing organs like lenses, how do plants work out the precise direction from which light is coming ?
In the absence of obvious physical sensing organs like lenses, how do plants work out the precise direction from which light is coming ?
Yet, studies show there’s only so much protein the body can use.

Yet, studies show there’s only so much protein the body can use.
𝐓𝐡𝐞 𝐬𝐢𝐦𝐩𝐥𝐞𝐬𝐭 𝐞𝐱𝐩𝐥𝐚𝐧𝐚𝐭𝐢𝐨𝐧 𝐢𝐬𝐧’𝐭 𝐚𝐥𝐰𝐚𝐲𝐬 𝐭𝐡𝐞 𝐛𝐞𝐬𝐭 𝐨𝐧𝐞.
1/n

𝐓𝐡𝐞 𝐬𝐢𝐦𝐩𝐥𝐞𝐬𝐭 𝐞𝐱𝐩𝐥𝐚𝐧𝐚𝐭𝐢𝐨𝐧 𝐢𝐬𝐧’𝐭 𝐚𝐥𝐰𝐚𝐲𝐬 𝐭𝐡𝐞 𝐛𝐞𝐬𝐭 𝐨𝐧𝐞.
1/n
Even if spectacularly successful, they leave open key questions:
◾️Functions of genetic mutations & polymorphisms
◾️Proteoform functions
◾️Protein abundance regulation
Biology is complex.

Even if spectacularly successful, they leave open key questions:
◾️Functions of genetic mutations & polymorphisms
◾️Proteoform functions
◾️Protein abundance regulation
Biology is complex.
Each tile in the maps represents a gene product; its size is proportional to the fraction of the gene product in hepatocytes.

Each tile in the maps represents a gene product; its size is proportional to the fraction of the gene product in hepatocytes.
GAPDH is often slandered as a 'housekeeping' protein, and its abundance varies significantly across human tissues.
How do you define a 'housekeeping' protein ?

GAPDH is often slandered as a 'housekeeping' protein, and its abundance varies significantly across human tissues.
How do you define a 'housekeeping' protein ?
1/2

1/2
𝐄𝐯𝐚𝐥𝐮𝐚𝐭𝐢𝐧𝐠 𝐦𝐨𝐝𝐞𝐥𝐬 𝐢𝐬 𝐡𝐚𝐫𝐝. 𝐈𝐭 𝐧𝐞𝐞𝐝𝐬 𝐦𝐨𝐫𝐞 𝐚𝐭𝐭𝐞𝐧𝐭𝐢𝐨𝐧 𝐚𝐧𝐝 𝐫𝐞𝐬𝐨𝐮𝐫𝐜𝐞𝐬.
1/2

𝐄𝐯𝐚𝐥𝐮𝐚𝐭𝐢𝐧𝐠 𝐦𝐨𝐝𝐞𝐥𝐬 𝐢𝐬 𝐡𝐚𝐫𝐝. 𝐈𝐭 𝐧𝐞𝐞𝐝𝐬 𝐦𝐨𝐫𝐞 𝐚𝐭𝐭𝐞𝐧𝐭𝐢𝐨𝐧 𝐚𝐧𝐝 𝐫𝐞𝐬𝐨𝐮𝐫𝐜𝐞𝐬.
1/2
We can start analyzing it quantitatively at higher resolution & scale.
A promising new direction for aging research 🚀
We find that the protein changes are cell-type specific, and thus muted in previous bulk analysis.
Protein clearance rates change with age and drive the ...
www.biorxiv.org/content/10.1...

We can start analyzing it quantitatively at higher resolution & scale.
A promising new direction for aging research 🚀
Had fun writing this, ping me if interested in other search engines. #TeamMassSpec
www.parallelsq.org/blog/fantast...

Had fun writing this, ping me if interested in other search engines. #TeamMassSpec
www.parallelsq.org/blog/fantast...
Peptides from some proteoforms correlate better to epitope-based measurements (Olink) than others.
The measurements differ because they reflect different proteoforms !

Peptides from some proteoforms correlate better to epitope-based measurements (Olink) than others.
The measurements differ because they reflect different proteoforms !
In the context of interactome networks, alternative isoforms tend to behave like distinct proteins rather than minor variants of each other.
This challenges:
1⃣ The interpretation of data from affinity reagents directed towards shared epitopes.
2⃣ The assignment of functions to genes.
When will biological research focus on such functional differences?

In the context of interactome networks, alternative isoforms tend to behave like distinct proteins rather than minor variants of each other.
This challenges:
1⃣ The interpretation of data from affinity reagents directed towards shared epitopes.
2⃣ The assignment of functions to genes.
When will biological research focus on such functional differences?

This challenges:
1⃣ The interpretation of data from affinity reagents directed towards shared epitopes.
2⃣ The assignment of functions to genes.
When will biological research focus on such functional differences?

