Siyu He
@siyuhe.bsky.social
Postdoc@stanford | PhD@columbia | AI4Biomedicine | Spatial biology | computational cancer biology | machine learning
Applying #CORAL on paired CODEX and Visium data from HCC, we delineated tissue ecosystems and identified macrophages interacting with CD4+ T cells as key players hindering responses.
For details, check our preprint & GitHub!😀
#Immunotherapy #HCC #SpatialOmics #CancerResearch
For details, check our preprint & GitHub!😀
#Immunotherapy #HCC #SpatialOmics #CancerResearch
February 8, 2025 at 4:00 PM
Applying #CORAL on paired CODEX and Visium data from HCC, we delineated tissue ecosystems and identified macrophages interacting with CD4+ T cells as key players hindering responses.
For details, check our preprint & GitHub!😀
#Immunotherapy #HCC #SpatialOmics #CancerResearch
For details, check our preprint & GitHub!😀
#Immunotherapy #HCC #SpatialOmics #CancerResearch
#CORAL enables the prediction of cell-cell interactions, as demonstrated in the mouse thymus. The latent variables in the model capture spatial features that transition from the medulla to the cortex, associating with thymic development and T cell maturation. 🧬5/🪸

February 8, 2025 at 4:00 PM
#CORAL enables the prediction of cell-cell interactions, as demonstrated in the mouse thymus. The latent variables in the model capture spatial features that transition from the medulla to the cortex, associating with thymic development and T cell maturation. 🧬5/🪸
Given paired spatial proteomics and downsampled ST data (such as Visium), #CORAL performs comparably to #SpatialGlue in detecting functional domains, while also achieving high-res ST🔬4/🪸

February 8, 2025 at 4:00 PM
Given paired spatial proteomics and downsampled ST data (such as Visium), #CORAL performs comparably to #SpatialGlue in detecting functional domains, while also achieving high-res ST🔬4/🪸
By leveraging deep generative model and graph attention mechanisms, #CORAL offers key benefits:
🔹 Spatial domain detection
🔹 Single-cell modality imputation
🔹 Inferring cell-cell interactions
Unlocking new possibilities in analyzing #spatialmultiomics integration! 3/🪸
🔹 Spatial domain detection
🔹 Single-cell modality imputation
🔹 Inferring cell-cell interactions
Unlocking new possibilities in analyzing #spatialmultiomics integration! 3/🪸
February 8, 2025 at 4:00 PM
By leveraging deep generative model and graph attention mechanisms, #CORAL offers key benefits:
🔹 Spatial domain detection
🔹 Single-cell modality imputation
🔹 Inferring cell-cell interactions
Unlocking new possibilities in analyzing #spatialmultiomics integration! 3/🪸
🔹 Spatial domain detection
🔹 Single-cell modality imputation
🔹 Inferring cell-cell interactions
Unlocking new possibilities in analyzing #spatialmultiomics integration! 3/🪸
Big thanks to our collaborators Matthew Bieniosek,
SongDongyuan,zhou_jingtian, benchidester, ZhenqinWu, Joseph Boen, Padmanee Sharma! And a special thanks to my advisors @jameszou.bsky.social and Alex Trevino for their invaluable guidance and support. 2/🪸
SongDongyuan,zhou_jingtian, benchidester, ZhenqinWu, Joseph Boen, Padmanee Sharma! And a special thanks to my advisors @jameszou.bsky.social and Alex Trevino for their invaluable guidance and support. 2/🪸
February 8, 2025 at 3:59 PM
Big thanks to our collaborators Matthew Bieniosek,
SongDongyuan,zhou_jingtian, benchidester, ZhenqinWu, Joseph Boen, Padmanee Sharma! And a special thanks to my advisors @jameszou.bsky.social and Alex Trevino for their invaluable guidance and support. 2/🪸
SongDongyuan,zhou_jingtian, benchidester, ZhenqinWu, Joseph Boen, Padmanee Sharma! And a special thanks to my advisors @jameszou.bsky.social and Alex Trevino for their invaluable guidance and support. 2/🪸
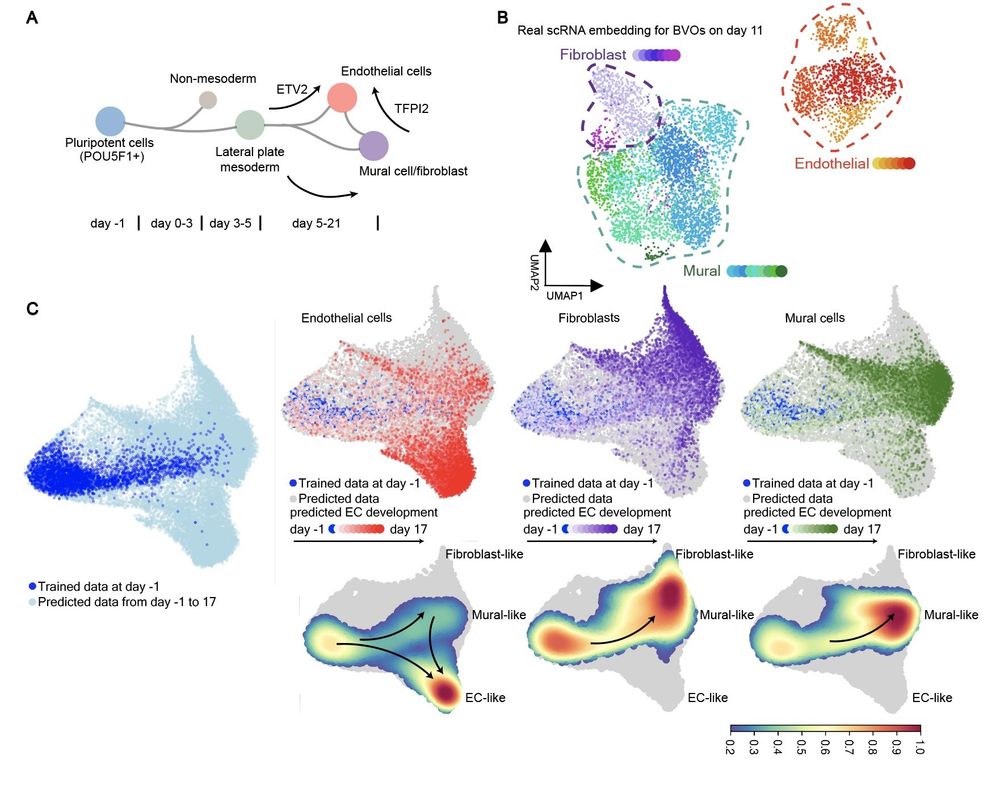
We applied Squidiff to model blood vessel #organoids using single-cell RNA sequencing to examine their response to neutron irradiation and pro-regenerative G-CSF treatment, simulating cosmic irradiation anticipated in #deepspace missions🧑🚀. 5/🦑
December 9, 2024 at 2:13 PM
We applied Squidiff to model blood vessel #organoids using single-cell RNA sequencing to examine their response to neutron irradiation and pro-regenerative G-CSF treatment, simulating cosmic irradiation anticipated in #deepspace missions🧑🚀. 5/🦑
Squidiff effectively predicts non-additive gene perturbation and cell type-specific drug responses, as demonstrated in its application to the analysis of temporal cell states in the glioblastoma dataset in response to novel drug combinations. 4/🦑
December 9, 2024 at 2:13 PM
Squidiff effectively predicts non-additive gene perturbation and cell type-specific drug responses, as demonstrated in its application to the analysis of temporal cell states in the glioblastoma dataset in response to novel drug combinations. 4/🦑
Given the scRNA data on day 0 and day 3, can we predict the data on day 1, day 2 and further? Squidiff predicts this differentiation of induced pluripotent stem cells (iPSCs) into mesendoderm and endoderm, guided by stimuli vectors. 3/🦑

December 9, 2024 at 2:13 PM
Given the scRNA data on day 0 and day 3, can we predict the data on day 1, day 2 and further? Squidiff predicts this differentiation of induced pluripotent stem cells (iPSCs) into mesendoderm and endoderm, guided by stimuli vectors. 3/🦑
Thank you so much for the long-term collaboration: Yuefei, @naveedtavakol.bsky.social , Haotian, and Sima. A special thanks to my advisors, James Zou, @elhamazizi.bsky.social , and Kam W. Leong, whose guidance and support have been invaluable. 2/🦑
December 9, 2024 at 2:13 PM
Thank you so much for the long-term collaboration: Yuefei, @naveedtavakol.bsky.social , Haotian, and Sima. A special thanks to my advisors, James Zou, @elhamazizi.bsky.social , and Kam W. Leong, whose guidance and support have been invaluable. 2/🦑

