Siyu He
@siyuhe.bsky.social
Postdoc@stanford | PhD@columbia | AI4Biomedicine | Spatial biology | computational cancer biology | machine learning
Reposted by Siyu He
New paper out in @natmethods.nature.com from @elhamazizi.bsky.social, Kam Leong & @jameszou.bsky.social! The team developed Squidiff, a diffusion #AI model to predict cellular responses to environmental cues and accelerate #PrecisionMedicine.
Learn more: bit.ly/3WRPNsx
Learn more: bit.ly/3WRPNsx

November 10, 2025 at 1:59 PM
New paper out in @natmethods.nature.com from @elhamazizi.bsky.social, Kam Leong & @jameszou.bsky.social! The team developed Squidiff, a diffusion #AI model to predict cellular responses to environmental cues and accelerate #PrecisionMedicine.
Learn more: bit.ly/3WRPNsx
Learn more: bit.ly/3WRPNsx
Reposted by Siyu He
Missed last week’s #CompSpatialBio seminar @justjhong.bsky.social with @siyuhe.bsky.social on CORAL—a deep graph model for spatial-omics? The recording is now available: youtu.be/BV5soaUVUNA
📅 Stay tuned for the next session! csbseminar.github.io
📅 Stay tuned for the next session! csbseminar.github.io

Learning single-cell spatial context through integrated spatial multiomics with CORAL - Siyu He
YouTube video by Computational Spatial Biology seminar
youtu.be
May 13, 2025 at 2:54 PM
Missed last week’s #CompSpatialBio seminar @justjhong.bsky.social with @siyuhe.bsky.social on CORAL—a deep graph model for spatial-omics? The recording is now available: youtu.be/BV5soaUVUNA
📅 Stay tuned for the next session! csbseminar.github.io
📅 Stay tuned for the next session! csbseminar.github.io
Thanks @justjhong.bsky.social for the invitation! Looking forward to the discussion.
For our next Computational Spatial Biology seminar, we will talking with @siyuhe.bsky.social about CORAL, a deep graph generative model learning integrated representations of multimodal spatial omics data.
Tune in next Wednesday, April 30 @ 12PM ET!
Full schedule: csbseminar.github.io
Tune in next Wednesday, April 30 @ 12PM ET!
Full schedule: csbseminar.github.io

April 24, 2025 at 5:08 PM
Thanks @justjhong.bsky.social for the invitation! Looking forward to the discussion.
Reposted by Siyu He
1/10 Excited to share our latest - the first whole-body map of both DNA methylation and 3D genome at single-cell resolution.
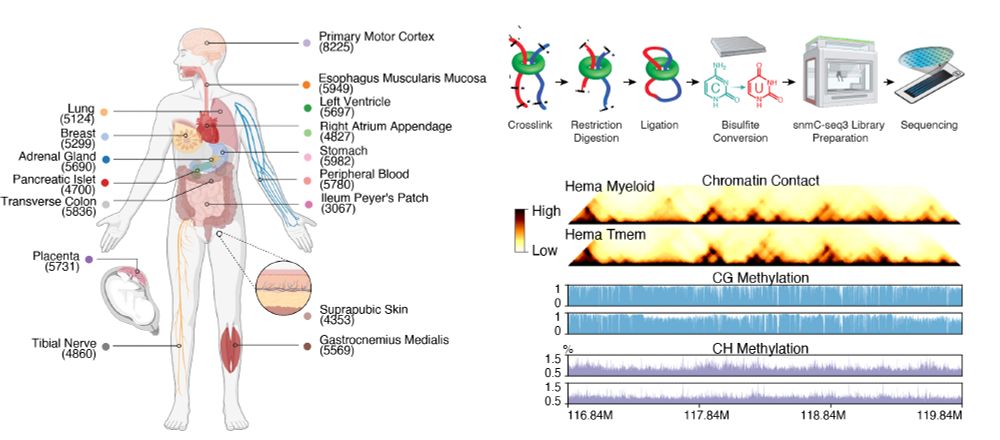
March 25, 2025 at 3:49 PM
1/10 Excited to share our latest - the first whole-body map of both DNA methylation and 3D genome at single-cell resolution.
Reposted by Siyu He
Check out our new collaborative work with Dr. Kam Leong @columbiauniversity.bsky.social now out in Advanced Functional Materials! We developed oral nanoparticles to help mitigate hematopoietic acute radiation syndrome, especially in cases of deep space exploration:
t.co/1jQwcU1pDB
t.co/1jQwcU1pDB

February 24, 2025 at 5:40 PM
Check out our new collaborative work with Dr. Kam Leong @columbiauniversity.bsky.social now out in Advanced Functional Materials! We developed oral nanoparticles to help mitigate hematopoietic acute radiation syndrome, especially in cases of deep space exploration:
t.co/1jQwcU1pDB
t.co/1jQwcU1pDB
Thrilled to introduce #CORAL🪸—a deep probabilistic graphical model that integrates spatial multiomics data!
🔹Resolves spatial resolution discrepancies across modalities
🔹Deconvolutes low-res data to achieve single-cell granularity
🔹Enables parallel integrative analysis 1/🪸
🔹Resolves spatial resolution discrepancies across modalities
🔹Deconvolutes low-res data to achieve single-cell granularity
🔹Enables parallel integrative analysis 1/🪸

February 8, 2025 at 3:58 PM
Thrilled to introduce #CORAL🪸—a deep probabilistic graphical model that integrates spatial multiomics data!
🔹Resolves spatial resolution discrepancies across modalities
🔹Deconvolutes low-res data to achieve single-cell granularity
🔹Enables parallel integrative analysis 1/🪸
🔹Resolves spatial resolution discrepancies across modalities
🔹Deconvolutes low-res data to achieve single-cell granularity
🔹Enables parallel integrative analysis 1/🪸
We are thrilled to share #Squidiff 🦑, a conditional diffusion model, which generates new transcriptomes that represent distinct cellular states, and its application to cell differentiation and drug perturbation www.biorxiv.org/content/10.1... 1/🦑

Squidiff: Predicting cellular development and responses to perturbations using a diffusion model
Single-cell sequencing has revolutionized our understanding of cellular heterogeneity and responses to environmental stimuli. However, mapping transcriptomic changes across diverse cell types in respo...
tinyurl.com
December 9, 2024 at 2:13 PM
We are thrilled to share #Squidiff 🦑, a conditional diffusion model, which generates new transcriptomes that represent distinct cellular states, and its application to cell differentiation and drug perturbation www.biorxiv.org/content/10.1... 1/🦑

