Researcher in infectious disease evolution
Group leader at Instiut Pasteur, Paris
https://research.pasteur.fr/en/member/sebastian-duchene-garzon/
Keen swimmer 🏊🏼♂️, runner 👟, and beach lover 🌊🤿🏖️
Views are my own
@pasteur.fr).
www.biorxiv.org/content/10.1...
Summary in replies (1/3)
@pasteur.fr).
www.biorxiv.org/content/10.1...
Summary in replies (1/3)
The Next Pandemic? A Scenario. open.substack.com/pub/markhoni...

The Next Pandemic? A Scenario. open.substack.com/pub/markhoni...
research.pasteur.fr/en/job/postd...

research.pasteur.fr/en/job/postd...
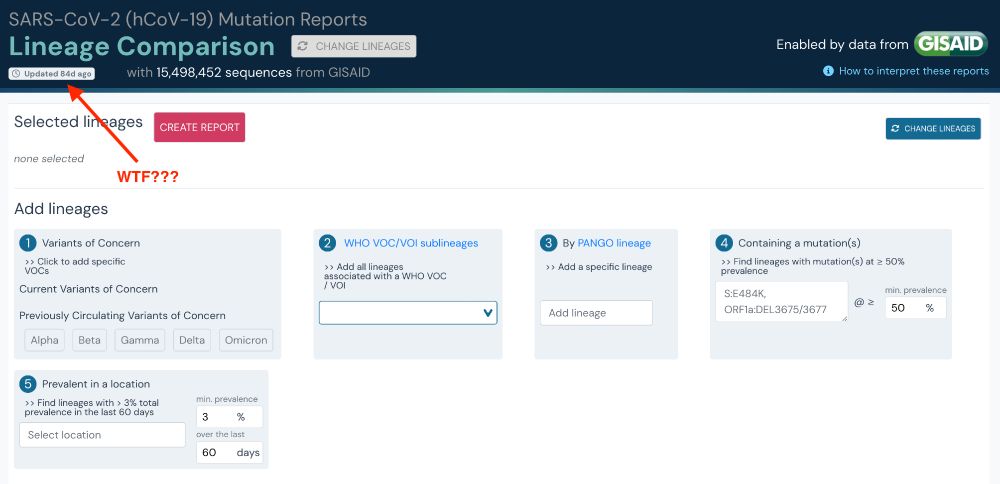
Well, the reason is that GISAID cut our access in January - as they did others. Without telling us.
After a ♾️ back and forth, that is now permanent.
More on that soon - and why GISAID should not be trusted with critical data.
Well, the reason is that GISAID cut our access in January - as they did others. Without telling us.
After a ♾️ back and forth, that is now permanent.
More on that soon - and why GISAID should not be trusted with critical data.