Come help us find new ways to understand and drug intrinsically disordered proteins (IDPs), it's a very interesting and important problem!
Link in the reply ↓
Come help us find new ways to understand and drug intrinsically disordered proteins (IDPs), it's a very interesting and important problem!
Link in the reply ↓

www.pnas.org/doi/10.1073/...
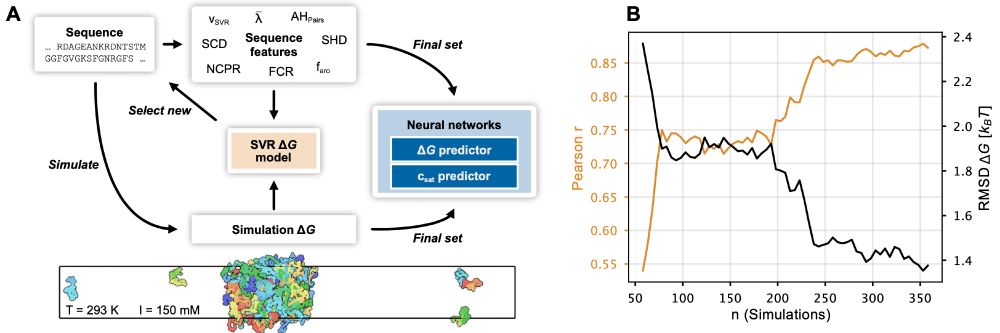
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n
www.pnas.org/doi/10.1073/...
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n

@bindresearch.org is hiring across the board: bindresearch.org/careers/
#compchem #compbio #NMRChat #TeamMassSpec #chemjobs @chemjobber.bsky.social
@bindresearch.org is hiring across the board: bindresearch.org/careers/
#compchem #compbio #NMRChat #TeamMassSpec #chemjobs @chemjobber.bsky.social
🤝 Hosted by Dr. @xsalvatella1.bsky.social
Register now! ➡️ shorturl.at/Q1LVA
#BarcelonaBiomed
🧪

🤝 Hosted by Dr. @xsalvatella1.bsky.social
Register now! ➡️ shorturl.at/Q1LVA
#BarcelonaBiomed
🧪

Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪

Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪

