Interested in ML for science/Compuational drug discovery/AI-assisted scientific discovery 🤞
from 🇱🇰🫶 https://ramith.fyi
bsky.app/profile/rami...

py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)

py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)
First up is Cielo, a Cray XE6 I worked on at LANL! Which might actually be the prettiest supercomputer I’ve worked on.

First up is Cielo, a Cray XE6 I worked on at LANL! Which might actually be the prettiest supercomputer I’ve worked on.
Forgot to change those settings, and the spark froze without me being able to ssh 😆
*In the spark, it’s unified mem
Gotto test again either reduced reallocation

Forgot to change those settings, and the spark froze without me being able to ssh 😆
*In the spark, it’s unified mem
Gotto test again either reduced reallocation
In 2025, 🥇 went to a @cmurobotics.bsky.social student. 🥈🥉 to @cmuengineering.bsky.social.
Register to compete in the 2026 #3MTCMU competition by 2/4. cmu.is/2026-3MT-Compete

In 2025, 🥇 went to a @cmurobotics.bsky.social student. 🥈🥉 to @cmuengineering.bsky.social.
Register to compete in the 2026 #3MTCMU competition by 2/4. cmu.is/2026-3MT-Compete

Thinking of polishing it up further:
current version: gist.github.com/ramithuh/aa6...

Thinking of polishing it up further:
current version: gist.github.com/ramithuh/aa6...
Mark Nepo

Mark Nepo
DGX Spark: ~1.3 it/s
L40: ~3.5 it/s
(same batch size used here)

DGX Spark: ~1.3 it/s
L40: ~3.5 it/s
(same batch size used here)
- transformer with Pair-Biased Multi-Head Attention
| Model | Layers | Parameters |
+---------+------+------------+
| Encoder | 12 | ~130M |
| Decoder | 12 | ~130M |
| Denoiser| 14 | ~160M |
+---------+------+-----------+
- transformer with Pair-Biased Multi-Head Attention
| Model | Layers | Parameters |
+---------+------+------------+
| Encoder | 12 | ~130M |
| Decoder | 12 | ~130M |
| Denoiser| 14 | ~160M |
+---------+------+-----------+
Pearl: arxiv.org/abs/2510.24670
ODesign: odesign1.github.io/static/pdfs/...
OpenFold (published earlier this week)
Find the full list at:
#ProteinFolding #AlphafoldandCo bit.ly/biofold

Pearl: arxiv.org/abs/2510.24670
ODesign: odesign1.github.io/static/pdfs/...
OpenFold (published earlier this week)
Find the full list at:
#ProteinFolding #AlphafoldandCo bit.ly/biofold
I used to present like this, thinking that I was being "academic", "organized", and "professional".
BUT, from the audience's viewpoints, this sucks. 😱
Look how far they need to hold a long-term context to just make sense of what you're saying!
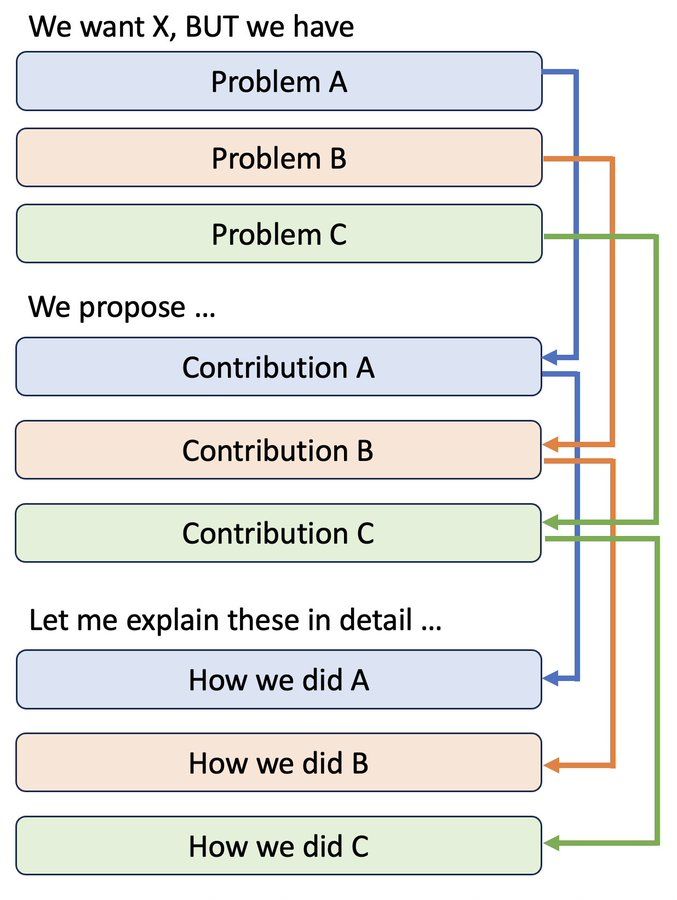
I used to present like this, thinking that I was being "academic", "organized", and "professional".
BUT, from the audience's viewpoints, this sucks. 😱
Look how far they need to hold a long-term context to just make sense of what you're saying!
Introducing: py2Dmol 🧬
(feedback, suggestions, requests are welcome)

Introducing: py2Dmol 🧬
(feedback, suggestions, requests are welcome)
The idea is to do prototyping in a cluster-agnostic manner (local GPU), then once the prototype is ready for a production run, use a dashboard to see which kinds of GPUs are available and submit it..
The idea is to do prototyping in a cluster-agnostic manner (local GPU), then once the prototype is ready for a production run, use a dashboard to see which kinds of GPUs are available and submit it..

*for a HW that's due midnight

*for a HW that's due midnight
back in 2014, i emailed a PM @ Intel with a suggestion, she kindly acknowleged it.. and the next day she asked for my address so that she could send me an intel galileo board..
(1/4)

back in 2014, i emailed a PM @ Intel with a suggestion, she kindly acknowleged it.. and the next day she asked for my address so that she could send me an intel galileo board..
(1/4)



