
@JLUGiessen, WGS bacteria, plasmids, software/pipeline developer, father of 2, husband, astrophotographer
A huge shout out and thank you to all Bakta users, bug reporters, those sharing ideas and suggesting features...
...just the entire incredibly supporting binfie community!
Without you, Bakta wouldn't be the same.
Thank you!

A huge shout out and thank you to all Bakta users, bug reporters, those sharing ideas and suggesting features...
...just the entire incredibly supporting binfie community!
Without you, Bakta wouldn't be the same.
Thank you!
Hundreds of thousands, even millions?
All annotated, taxonomically classified, integrated with metadata.
Easily searchable, viewable, downloadable, in sync with #AllTheBacteria.
Then BakRep is for you! Poster P-CM-102 @vaam-microbes.bsky.social #VAAM25

Hundreds of thousands, even millions?
All annotated, taxonomically classified, integrated with metadata.
Easily searchable, viewable, downloadable, in sync with #AllTheBacteria.
Then BakRep is for you! Poster P-CM-102 @vaam-microbes.bsky.social #VAAM25
Though not being a captain, that's the post, ... just that ;-)
Though not being a captain, that's the post, ... just that ;-)
- MUCH faster annotations via optimized Kubernetes nodes and locally-attached databases
- Software-Update to v1.10.1
- Database-Update to v5.1
Publicly available at: bakta.computational.bio
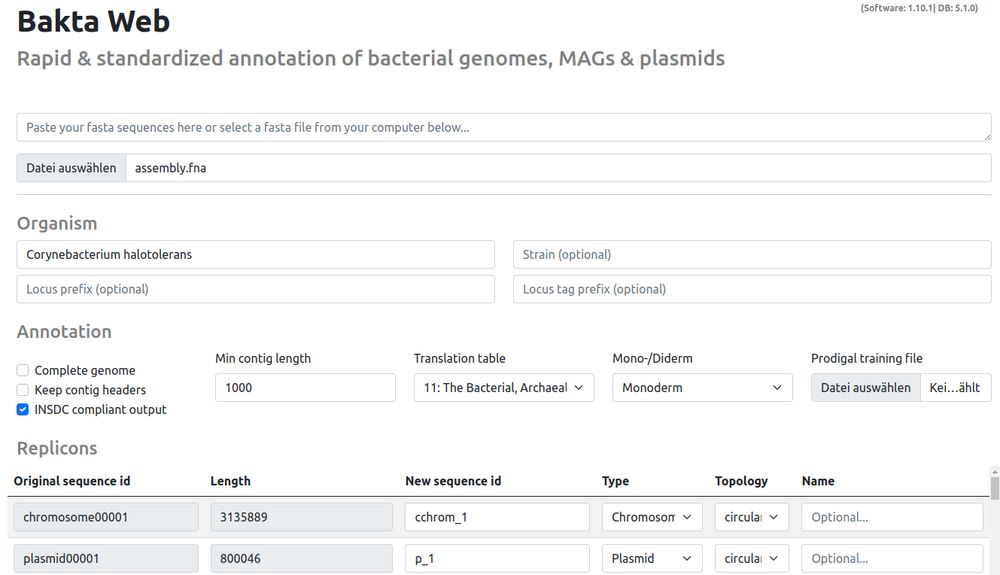
- MUCH faster annotations via optimized Kubernetes nodes and locally-attached databases
- Software-Update to v1.10.1
- Database-Update to v5.1
Publicly available at: bakta.computational.bio
To improve Bakta’s circular genome plots, we replaced good old Circos by the novel pure-Python pyCirclize library. Thus, we could also add new features, such as legends and new 'bakta_plot' parameters:
bakta_plot --type cog –dpi 300 --size 8 --label='my custom label text'
(6/11)
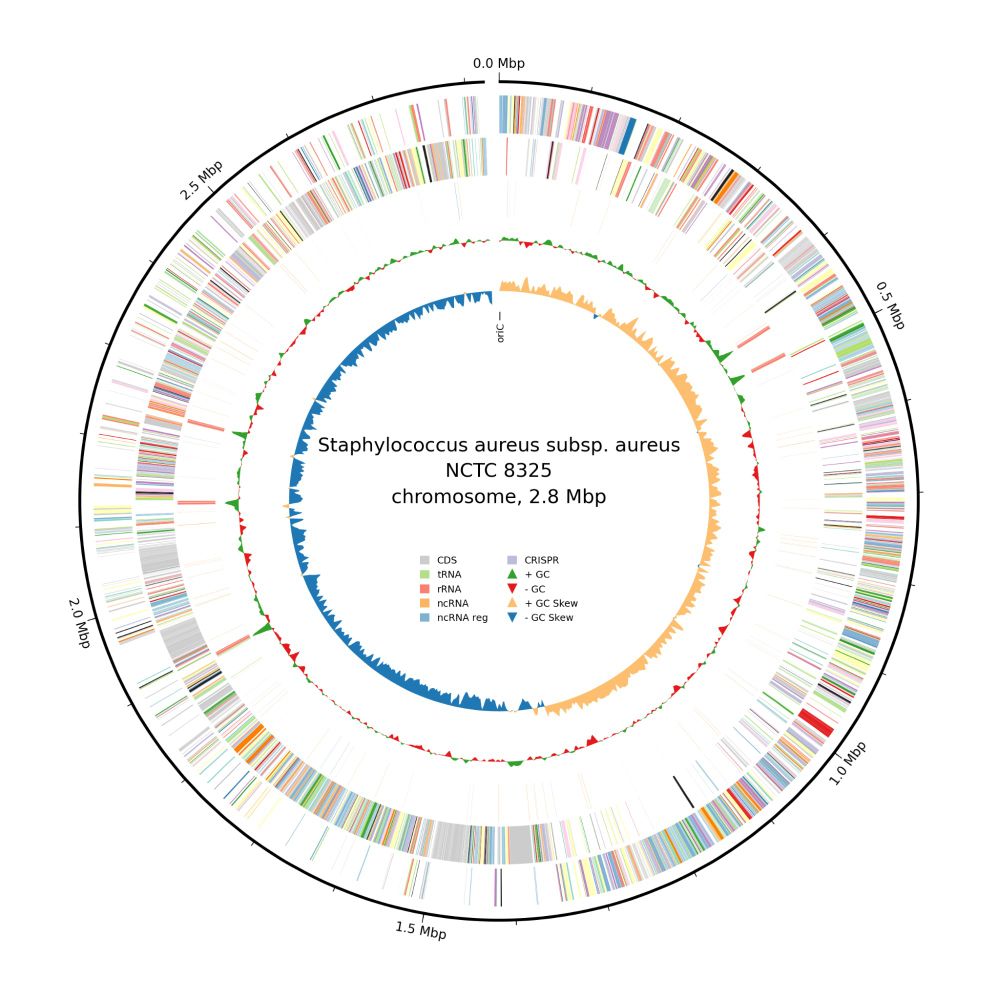
To improve Bakta’s circular genome plots, we replaced good old Circos by the novel pure-Python pyCirclize library. Thus, we could also add new features, such as legends and new 'bakta_plot' parameters:
bakta_plot --type cog –dpi 300 --size 8 --label='my custom label text'
(6/11)
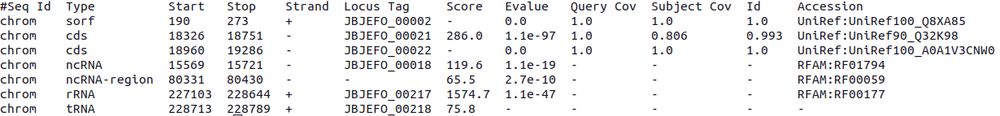
Bakta’s output too large? Via a new 'bakta_io' cmd all result files can be recovered from its json[.gz] file within seconds.
By this, ~84 Mb of result files (e.g. E.coli) are stored in a 5.8 Mb .json.gz file reducing storage requirements by 93%.
(3/11)
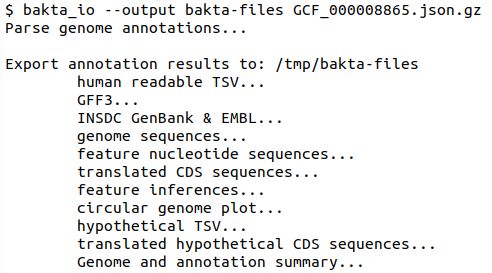
Bakta’s output too large? Via a new 'bakta_io' cmd all result files can be recovered from its json[.gz] file within seconds.
By this, ~84 Mb of result files (e.g. E.coli) are stored in a 5.8 Mb .json.gz file reducing storage requirements by 93%.
(3/11)
species=="Klebsiella pneumoniae"
+ contig number <= 100
+ completeness >= 0.95
+ contamination <= 0.01
+ containing gene=="mobA"
+ collection-host=="human"
-> 1,545 genomes in <2 sec 😀
5/7

species=="Klebsiella pneumoniae"
+ contig number <= 100
+ completeness >= 0.95
+ contamination <= 0.01
+ containing gene=="mobA"
+ collection-host=="human"
-> 1,545 genomes in <2 sec 😀
5/7

