
@JLUGiessen, WGS bacteria, plasmids, software/pipeline developer, father of 2, husband, astrophotographer

@MicrobioSoc
#MGen
>661,000 bacterial genomes, uniformly characterized & annotated, enriched with metadata, accessible via a flexible search engine:
doi.org/10.1099/mgen...
👇 1/7

We celebrated 10 years of de.NBI with a birthday cake 🎂, welcomed new Associated Partners, and connected with 140+ participants. Stay tuned for more! #deNBI2025




We celebrated 10 years of de.NBI with a birthday cake 🎂, welcomed new Associated Partners, and connected with 140+ participants. Stay tuned for more! #deNBI2025
A huge shout out and thank you to all Bakta users, bug reporters, those sharing ideas and suggesting features...
...just the entire incredibly supporting binfie community!
Without you, Bakta wouldn't be the same.
Thank you!

A huge shout out and thank you to all Bakta users, bug reporters, those sharing ideas and suggesting features...
...just the entire incredibly supporting binfie community!
Without you, Bakta wouldn't be the same.
Thank you!
To further improve the functional annotation of "hypothetical" CDS, me and @gbouras13.bsky.social, we are looking for the worst Bakta-annotated bacterial genomes ;-)
(1/2)
To further improve the functional annotation of "hypothetical" CDS, me and @gbouras13.bsky.social, we are looking for the worst Bakta-annotated bacterial genomes ;-)
(1/2)
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
3 sessions full of microbial bioinformatics at JLU Giessen:
I: QC & QA, assembly
II: regional & functional annotation
III: comparative genomics
Info & registration: www.denbi.de/training-cou...
3 sessions full of microbial bioinformatics at JLU Giessen:
I: QC & QA, assembly
II: regional & functional annotation
III: comparative genomics
Info & registration: www.denbi.de/training-cou...
doi.org/10.1093/nar/...
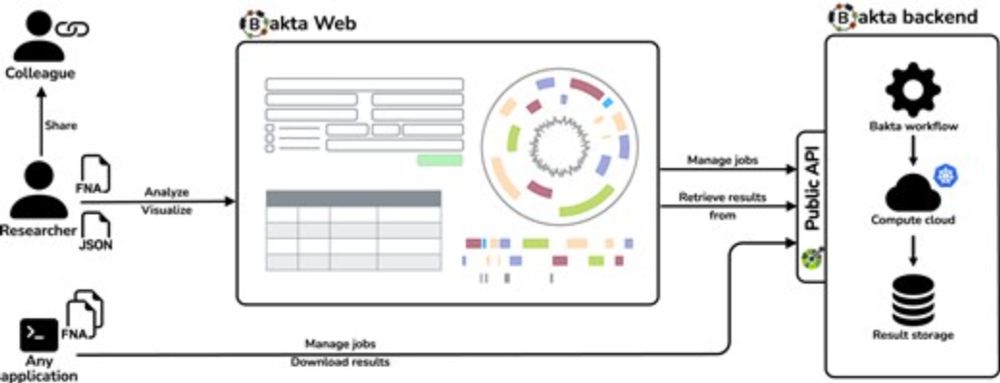
Easy to use, no registration, fast, scalable, various visualizations, in sync with Bakta CLI:
bakta.computational.bio
(1/5)
doi.org/10.1093/nar/...
Easy to use, no registration, fast, scalable, various visualizations, in sync with Bakta CLI:
bakta.computational.bio
(1/5)
Hundreds of thousands, even millions?
All annotated, taxonomically classified, integrated with metadata.
Easily searchable, viewable, downloadable, in sync with #AllTheBacteria.
Then BakRep is for you! Poster P-CM-102 @vaam-microbes.bsky.social #VAAM25

Hundreds of thousands, even millions?
All annotated, taxonomically classified, integrated with metadata.
Easily searchable, viewable, downloadable, in sync with #AllTheBacteria.
Then BakRep is for you! Poster P-CM-102 @vaam-microbes.bsky.social #VAAM25
After a year, it was time for a Bakta database update - and it's a huge one:
- IPS: 330.9M
- PSC: 135.3M
- PSCC: 37M
doi.org/10.5281/zeno...
👇 1/6
After a year, it was time for a Bakta database update - and it's a huge one:
- IPS: 330.9M
- PSC: 135.3M
- PSCC: 37M
doi.org/10.5281/zeno...
👇 1/6
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...
Though not being a captain, that's the post, ... just that ;-)
Though not being a captain, that's the post, ... just that ;-)
github.com/kjolley/BIGS...
github.com/kjolley/BIGS...
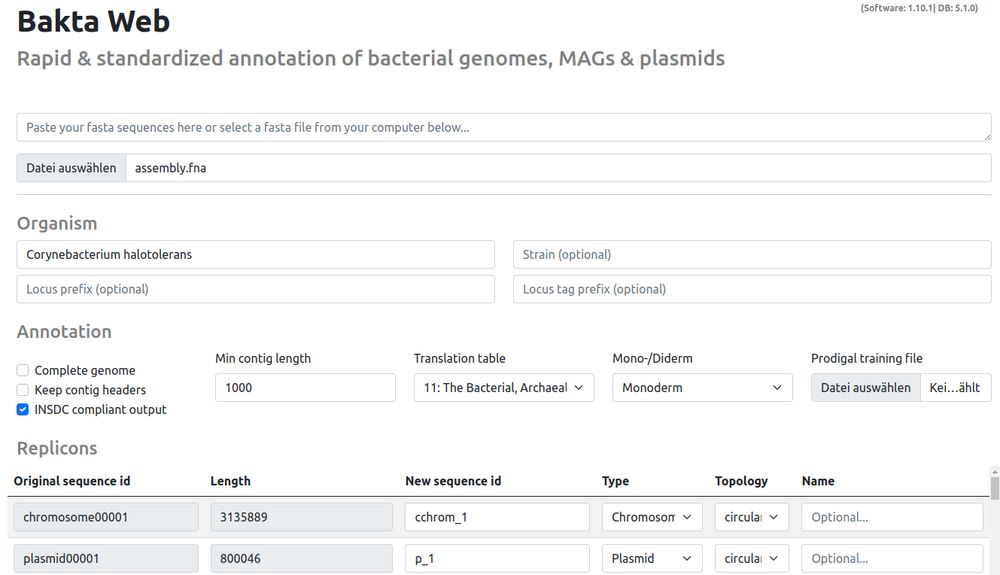
- MUCH faster annotations via optimized Kubernetes nodes and locally-attached databases
- Software-Update to v1.10.1
- Database-Update to v5.1
Publicly available at: bakta.computational.bio
- MUCH faster annotations via optimized Kubernetes nodes and locally-attached databases
- Software-Update to v1.10.1
- Database-Update to v5.1
Publicly available at: bakta.computational.bio
Highlights:
- user-provided HMMs: --hmms
- output file recovery from JSON files: bakta_io
- export of inference metrics: inference.tsv
- bypass overlap filters: --skip-filter
- improved genome plots
github.com/oschwengers/...
👇 (1/11)

Highlights:
- user-provided HMMs: --hmms
- output file recovery from JSON files: bakta_io
- export of inference metrics: inference.tsv
- bypass overlap filters: --skip-filter
- improved genome plots
github.com/oschwengers/...
👇 (1/11)
@MicrobioSoc
#MGen
>661,000 bacterial genomes, uniformly characterized & annotated, enriched with metadata, accessible via a flexible search engine:
doi.org/10.1099/mgen...
👇 1/7

@MicrobioSoc
#MGen
>661,000 bacterial genomes, uniformly characterized & annotated, enriched with metadata, accessible via a flexible search engine:
doi.org/10.1099/mgen...
👇 1/7
- bacteria: 30,238 -> 50,226
- archaea: 606 -> 905
- fungi: 347 -> 557
- plasmids: 32,611 -> 81,674
doi.org/10.5281/zeno...
- bacteria: 30,238 -> 50,226
- archaea: 606 -> 905
- fungi: 347 -> 557
- plasmids: 32,611 -> 81,674
doi.org/10.5281/zeno...
- new --regions option to provide pre-annotated feature regions
- annotation of spacer & repeat sequences in CRISPR arrays
github.com/oschwengers/...
More information below 👇 (1/7)
- new --regions option to provide pre-annotated feature regions
- annotation of spacer & repeat sequences in CRISPR arrays
github.com/oschwengers/...
More information below 👇 (1/7)

