Interested in microbial genomics, pangenome graphs & evolution 🧬🦠💻
This means all public tools/webapps of GISAID data (all the ones you've been used to seeing thru the pandemic, as far as we can tell) are prohibited.
The file allowed this. Cut that - cut off all tools the public & others were using.

This means all public tools/webapps of GISAID data (all the ones you've been used to seeing thru the pandemic, as far as we can tell) are prohibited.
The file allowed this. Cut that - cut off all tools the public & others were using.
Thread 1/n

Thread 1/n
Our group uses theoretical physics to understand how cells collectively self-organize.
If you're interested, get in touch & check out the Biozentrum PhD Fellowship program, deadline October 12th!

Our group uses theoretical physics to understand how cells collectively self-organize.
If you're interested, get in touch & check out the Biozentrum PhD Fellowship program, deadline October 12th!
We analyzed data collected by the fantastic SEA-PHAGES program in phagesdb.org.
[1/6]
www.biorxiv.org/content/10.1...
We analyzed data collected by the fantastic SEA-PHAGES program in phagesdb.org.
[1/6]
www.biorxiv.org/content/10.1...
Are you excited by the idea of building global infrastructure to make pathogen sequencing more accessible, interpretable, and equitable? 🧑🏻💻🧬
My group at @swisstph.ch has an opening working with ARTIC2, @pathoplexus.org, & Loculus - read on!
1/5

Are you excited by the idea of building global infrastructure to make pathogen sequencing more accessible, interpretable, and equitable? 🧑🏻💻🧬
My group at @swisstph.ch has an opening working with ARTIC2, @pathoplexus.org, & Loculus - read on!
1/5

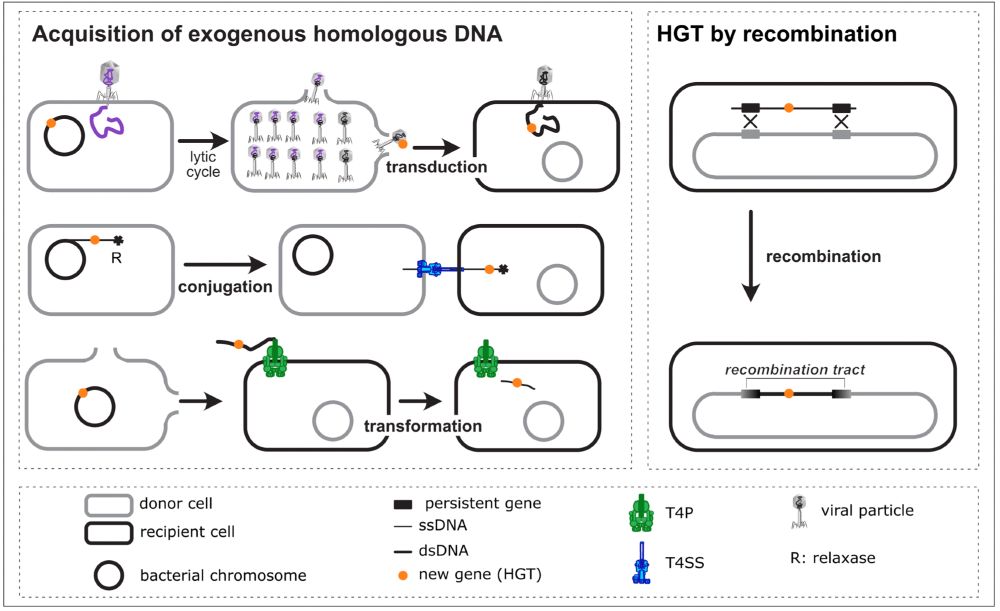
[1/N]
www.biorxiv.org/content/10.1...

[1/N]
www.biorxiv.org/content/10.1...
academic.oup.com/mbe/advance-...
Are you curious about how fast the genome of E.coli evolves structurally (gains, rearrangements...) ? 🧬
A summary thread [1/N]🧵

academic.oup.com/mbe/advance-...
Are you curious about how fast the genome of E.coli evolves structurally (gains, rearrangements...) ? 🧬
A summary thread [1/N]🧵
@pathoplexus.org ! 🙏
I'm proud to see Pathoplexus continuing to draw attention - but this is truly a team effort and only possible thanks to all the amazing people involved! 🙌
swisstph.ch/en/news/news...
1/2

@pathoplexus.org ! 🙏
I'm proud to see Pathoplexus continuing to draw attention - but this is truly a team effort and only possible thanks to all the amazing people involved! 🙌
swisstph.ch/en/news/news...
1/2
Though not everyone in Pathoplexus could be there in person, it was a fantastic & thought-provoking event, & I'm so glad were part of it! #OpenScience #OpenSource

Though not everyone in Pathoplexus could be there in person, it was a fantastic & thought-provoking event, & I'm so glad were part of it! #OpenScience #OpenSource

