Protein ML person, mathematics & science enthusiast.
developer of salad
preprint: https://www.biorxiv.org/content/10.1101/2025.01.31.635780v1
code: https://github.com/mjendrusch/salad
5/5
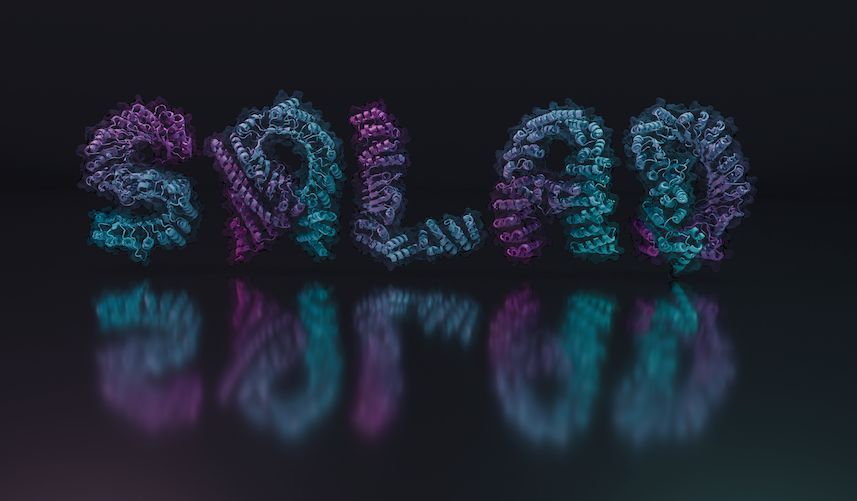
5/5
3/🧵


3/🧵
colab.research.google.com/github/mjend...
More improvements coming soon!

colab.research.google.com/github/mjend...
More improvements coming soon!

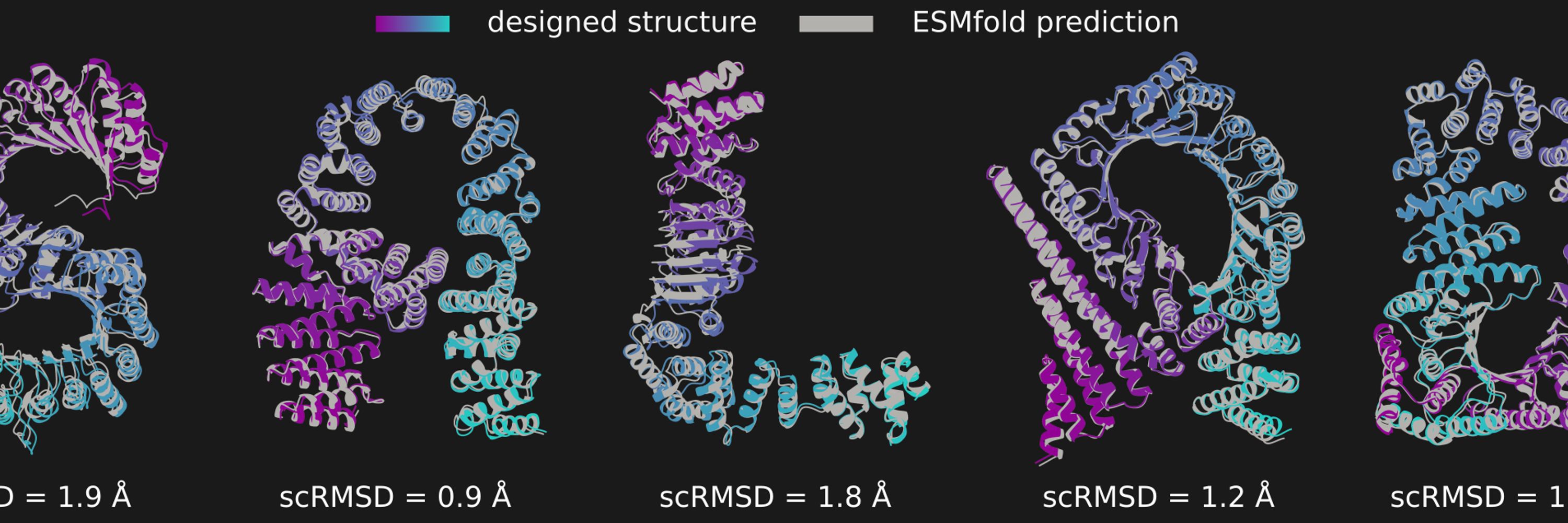
For smaller proteins, salad generations can get pretty crazy, though (400aa, upper row is the first 4 salad VP-scaled examples, lower row is the first 4 RSO examples I could find)

For smaller proteins, salad generations can get pretty crazy, though (400aa, upper row is the first 4 salad VP-scaled examples, lower row is the first 4 RSO examples I could find)
In the image you can see salad-generated structures spelling the word "SALAD", overlaid with their ESMfold predictions and scRMSD.
(6/N)
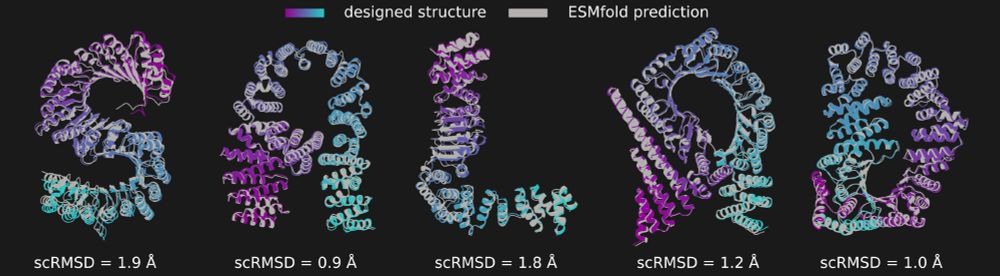
In the image you can see salad-generated structures spelling the word "SALAD", overlaid with their ESMfold predictions and scRMSD.
(6/N)
(5/N)



(5/N)
We extend RFdiffusion's sampling trick for symmetry to arbitrary edits to the input and output of salad models at each generation step.
This way, we can make salad work on design tasks it was not trained for ...
(4/N)

We extend RFdiffusion's sampling trick for symmetry to arbitrary edits to the input and output of salad models at each generation step.
This way, we can make salad work on design tasks it was not trained for ...
(4/N)
For proteins of this size one would otherwise have to use RSO or af2cycler so far – protein structure diffusion models do not work well.
Our best salad models (blue) bridge this designability gap to RSO.
(3/N)

For proteins of this size one would otherwise have to use RSO or af2cycler so far – protein structure diffusion models do not work well.
Our best salad models (blue) bridge this designability gap to RSO.
(3/N)
(and Genie 2 and Chroma and Proteus and many others)
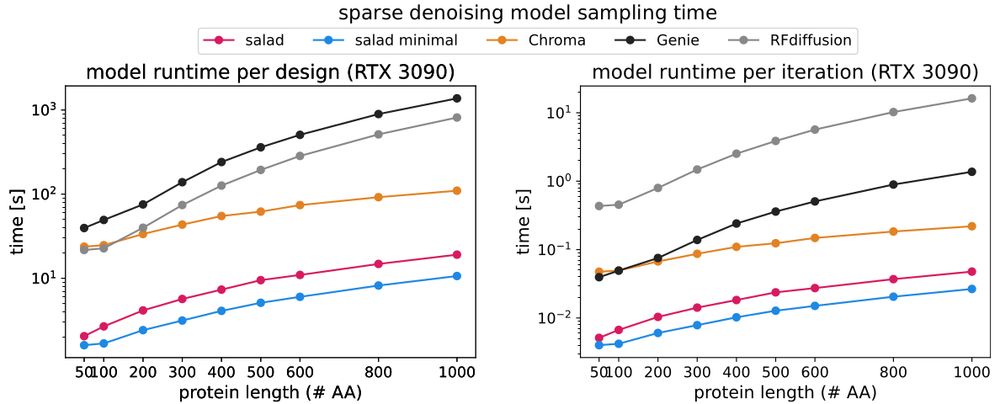
1. salad is tiny, with the denoising model clocking in at about 8.4 M parameters.
2. salad is fast, up to 42x faster than RFdiffusion on large proteins
(2/N)
(and Genie 2 and Chroma and Proteus and many others)
1. salad is tiny, with the denoising model clocking in at about 8.4 M parameters.
2. salad is fast, up to 42x faster than RFdiffusion on large proteins
(2/N)
We describe salad (sparse all-atom denoising), a family of efficient protein structure diffusion models and show that it works well on a bunch of protein design task previously described in the literature.
Preprint: www.biorxiv.org/content/10.1...
(1/N)

We describe salad (sparse all-atom denoising), a family of efficient protein structure diffusion models and show that it works well on a bunch of protein design task previously described in the literature.
Preprint: www.biorxiv.org/content/10.1...
(1/N)
(now with 100% more structure predictions and plausibility metrics)

(now with 100% more structure predictions and plausibility metrics)
you can spell out random messages as protein structures.

you can spell out random messages as protein structures.

