We ran simulations to estimate EP search times based on 1) measured subdiffusion, 2) physical/genomic distance. Within 300nm (~<200 kb), a gene can find 1000s of enhancers in minutes. Such encounters can’t be selective → EP selectivity needs other mechanisms, e.g. biochemical
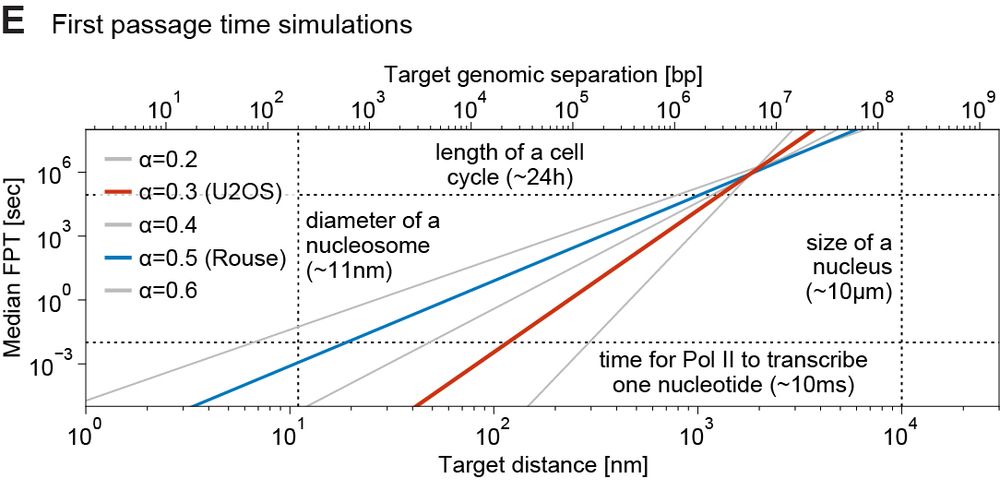
We ran simulations to estimate EP search times based on 1) measured subdiffusion, 2) physical/genomic distance. Within 300nm (~<200 kb), a gene can find 1000s of enhancers in minutes. Such encounters can’t be selective → EP selectivity needs other mechanisms, e.g. biochemical
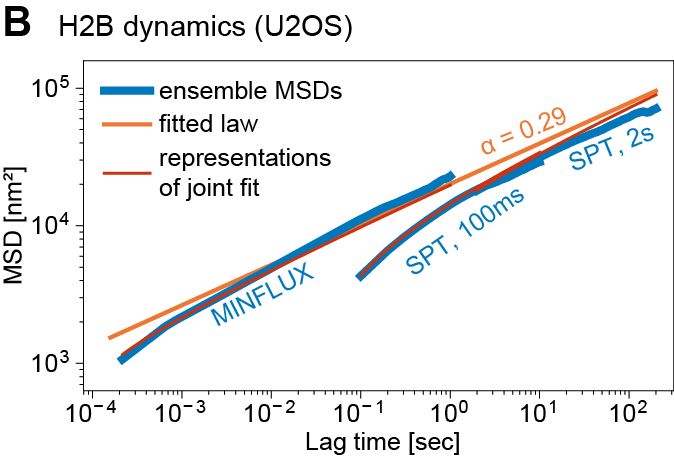
Such fast and long chromatin tracking is key to understanding how distal genomic elements (e.g. enhancers and promoters – EP) find each other. We found chromatin is highly subdiffusive (α ~ 0.3) -> loci are great at exploring the neighborhoods but rarely reach distant regions
Such fast and long chromatin tracking is key to understanding how distal genomic elements (e.g. enhancers and promoters – EP) find each other. We found chromatin is highly subdiffusive (α ~ 0.3) -> loci are great at exploring the neighborhoods but rarely reach distant regions
Super excited to share my first preprint from @andersshansen.bsky.social Lab! We used MINFLUX to track chromatin (H2B-Halo and Fbn2 locus) at an unprecedented 200 μs, then combined it with SPT to span μs-minutes (H2B) or SPT & Super-Res Live-Cell Imaging (SRLCI) to span μs-hours (Fbn2)
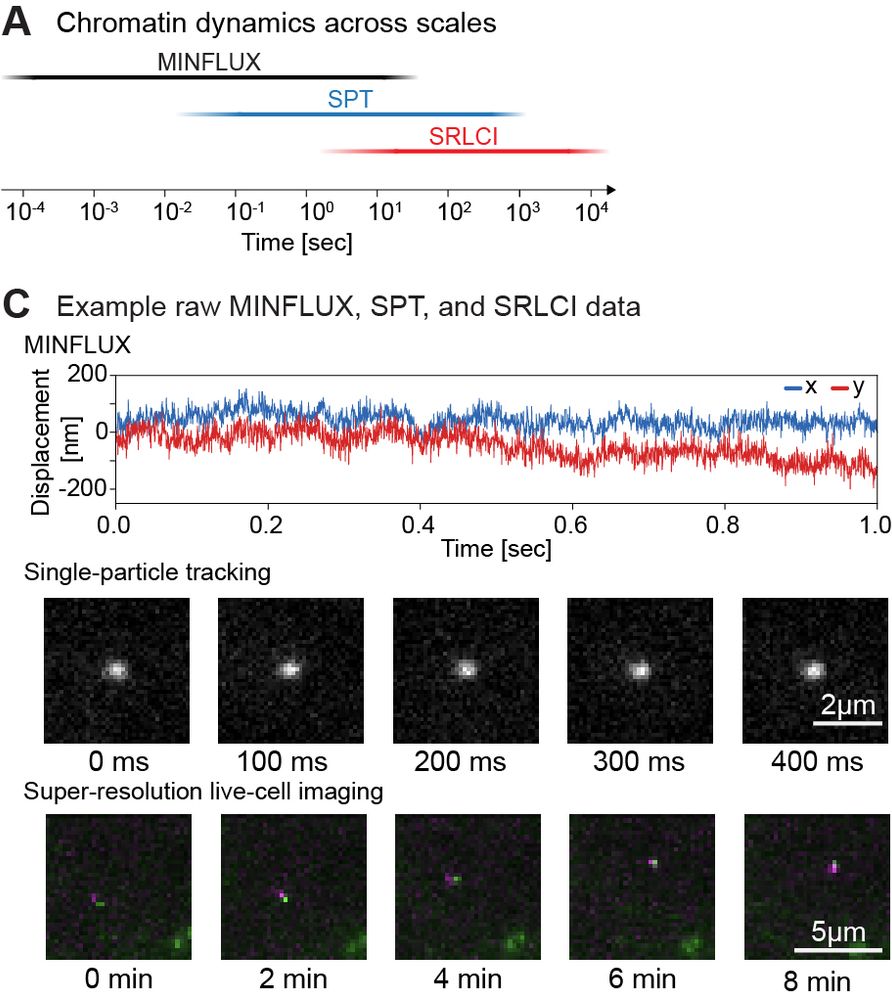
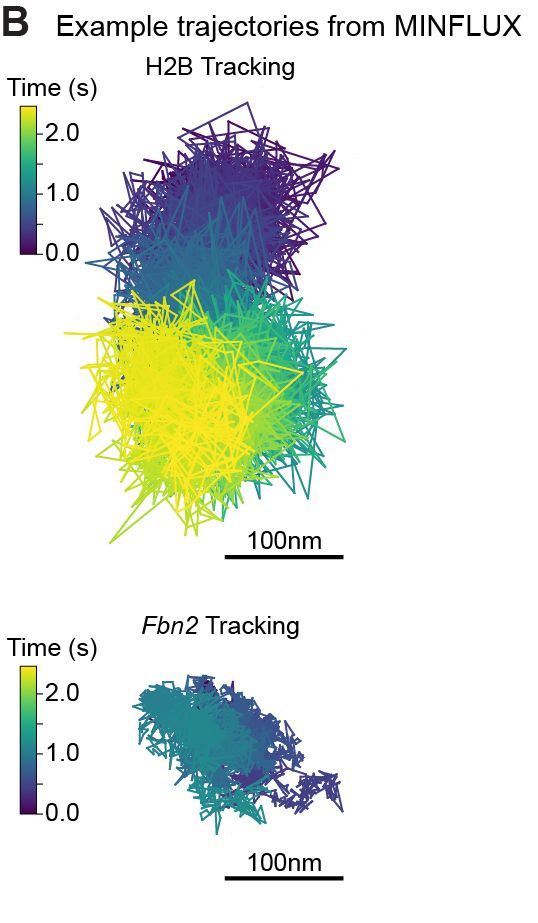
Super excited to share my first preprint from @andersshansen.bsky.social Lab! We used MINFLUX to track chromatin (H2B-Halo and Fbn2 locus) at an unprecedented 200 μs, then combined it with SPT to span μs-minutes (H2B) or SPT & Super-Res Live-Cell Imaging (SRLCI) to span μs-hours (Fbn2)

