🌐 evedesign.bio
🌐 evedesign.bio


- SIMD FW/BW alignment (preprint soon!)
- Sub. Mat. λ calculator by Eric Dawson
- Faster ARM SW by Alexander Nesterovskiy
- MSA-Pairformer’s proximity-based pairing for multimer prediction (www.biorxiv.org/content/10.1...; avail. in ColabFold API)
💾 github.com/soedinglab/M... & 🐍

- SIMD FW/BW alignment (preprint soon!)
- Sub. Mat. λ calculator by Eric Dawson
- Faster ARM SW by Alexander Nesterovskiy
- MSA-Pairformer’s proximity-based pairing for multimer prediction (www.biorxiv.org/content/10.1...; avail. in ColabFold API)
💾 github.com/soedinglab/M... & 🐍
- Added description texts for AFDB
- Integrated TaxoView taxonomy visualization & filter by @sunjaelee.bsky.social
- Inter-residue distance clustering by DBSCAN to explore motif diversity.
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
- Added description texts for AFDB
- Integrated TaxoView taxonomy visualization & filter by @sunjaelee.bsky.social
- Inter-residue distance clustering by DBSCAN to explore motif diversity.
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
📄 academic.oup.com/nar/article/...

📄 academic.oup.com/nar/article/...
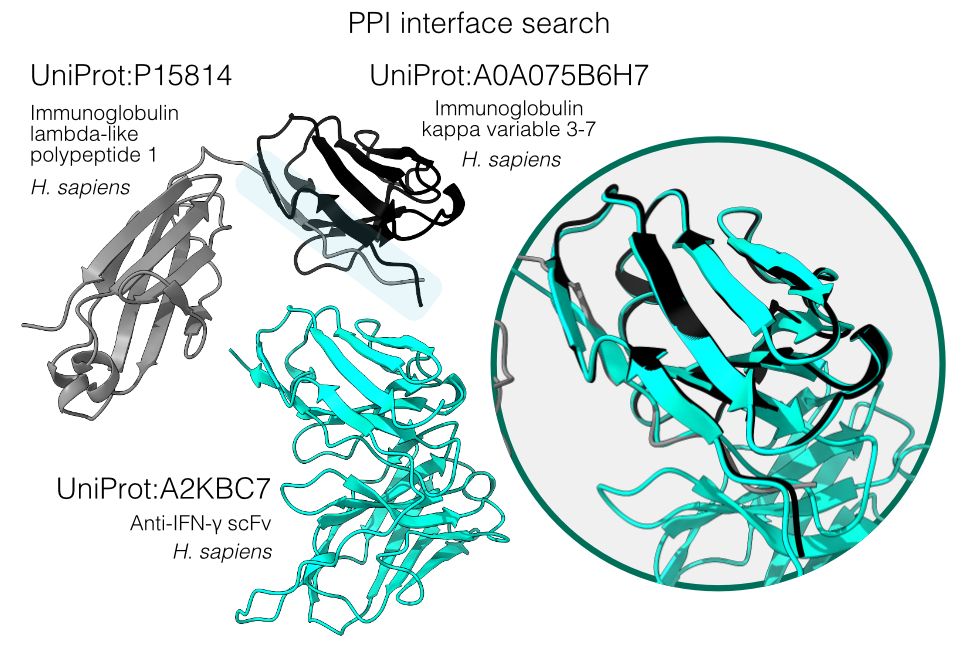
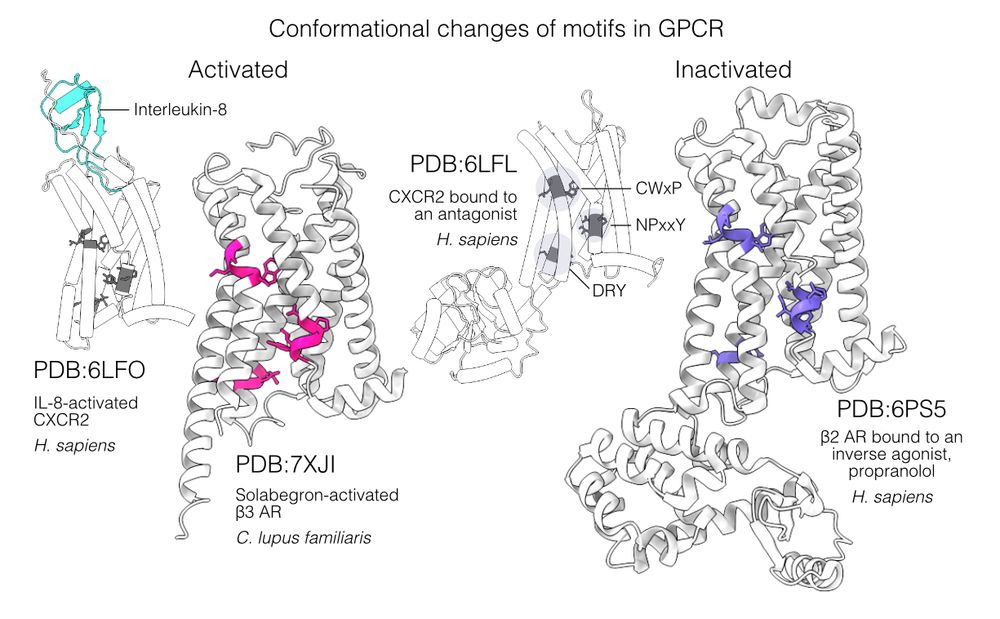
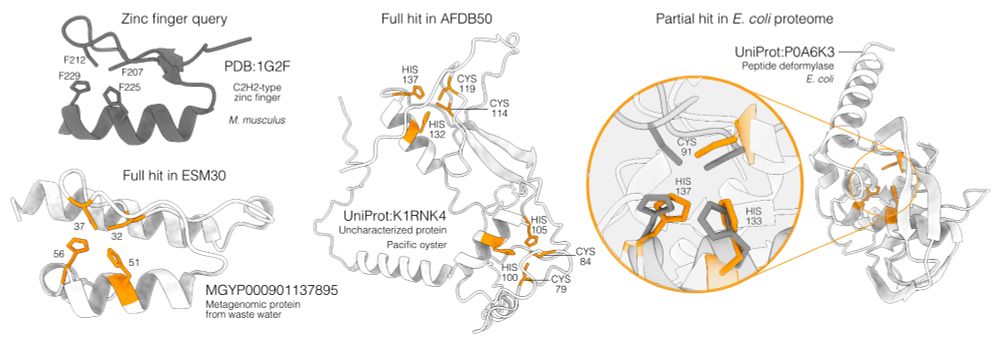
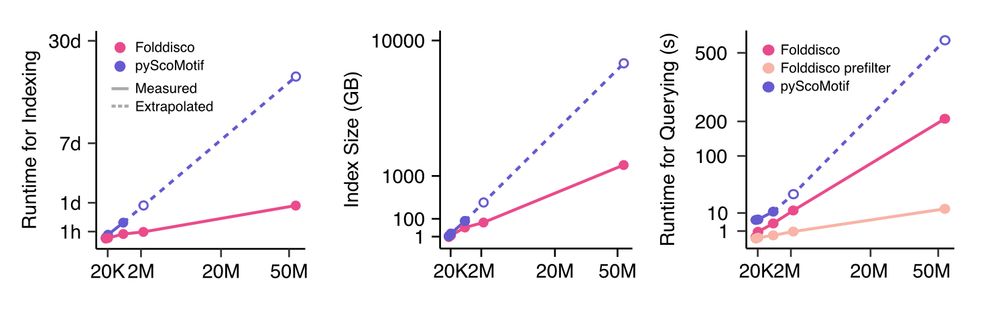
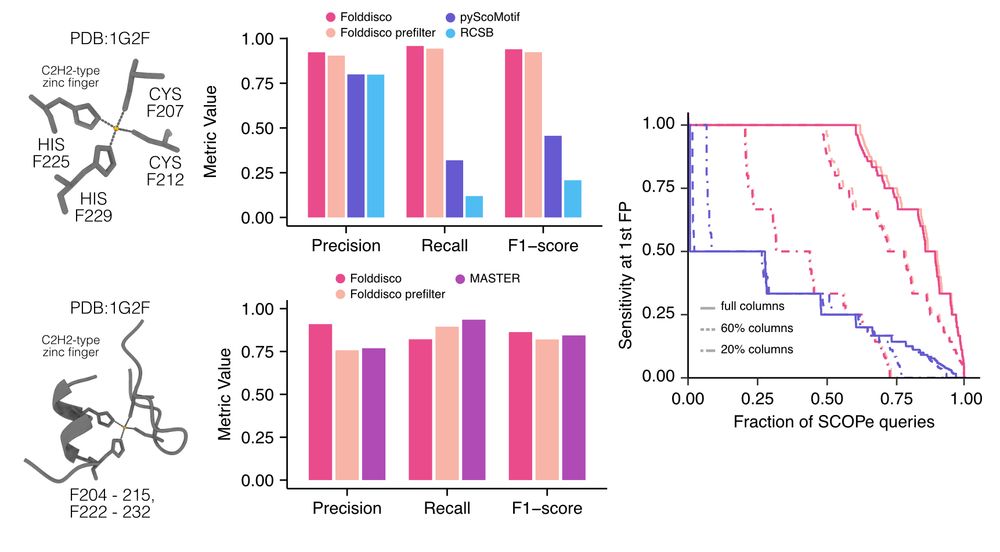
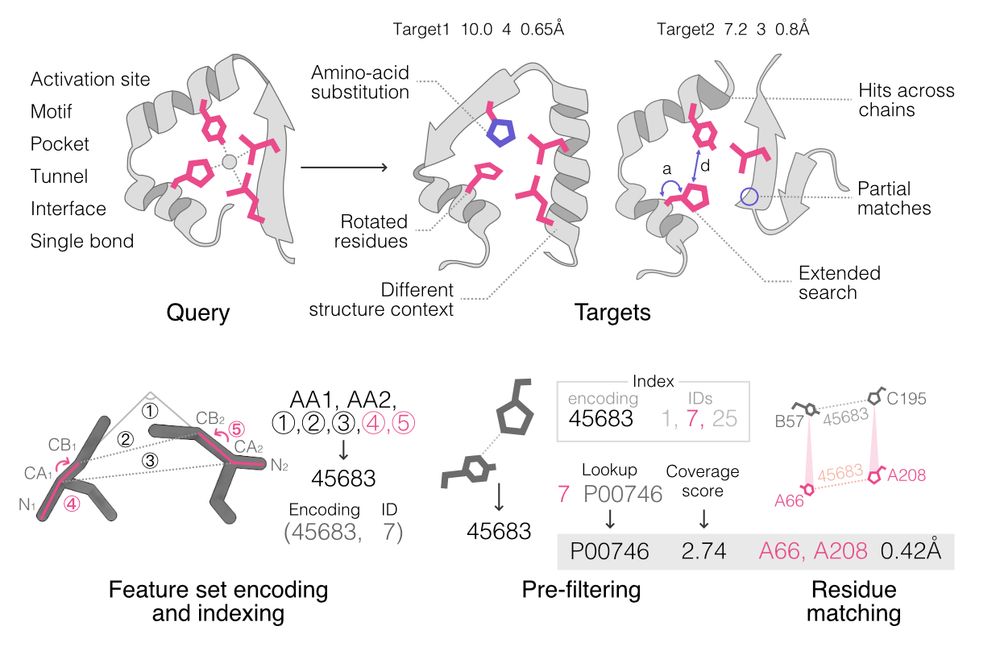
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
🌐 afesm.foldseek.com
Read more about the work in the skeetorial
🦋 bsky.app/profile/mart...
or our preprint
📄 www.biorxiv.org/content/10.1...
🌐 afesm.foldseek.com
Read more about the work in the skeetorial
🦋 bsky.app/profile/mart...
or our preprint
📄 www.biorxiv.org/content/10.1...



🌐 afesm.foldseek.com
🌐 afesm.foldseek.com
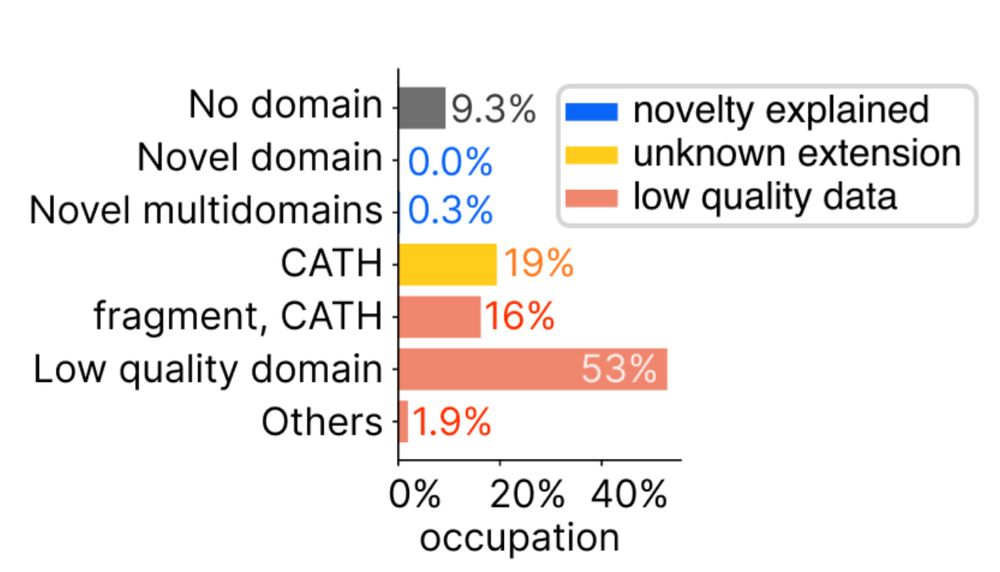
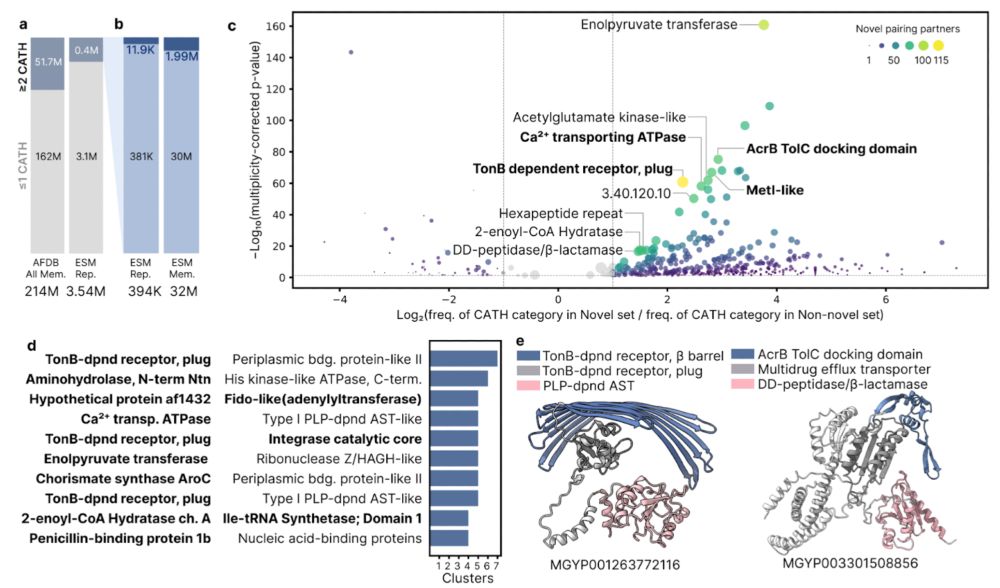
📄 doi.org/10.1126/scie... 4/n
📄 doi.org/10.1126/scie... 4/n