













www.nature.com/articles/s41...
www.nature.com/articles/s41...

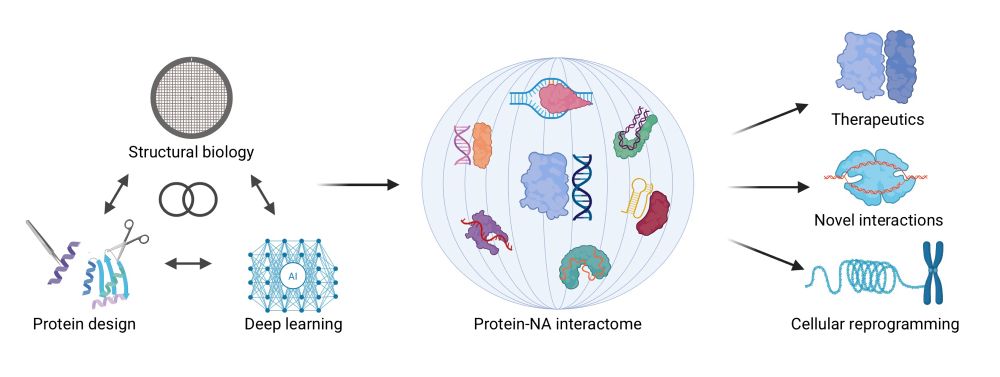


github.com/martinpacesa...
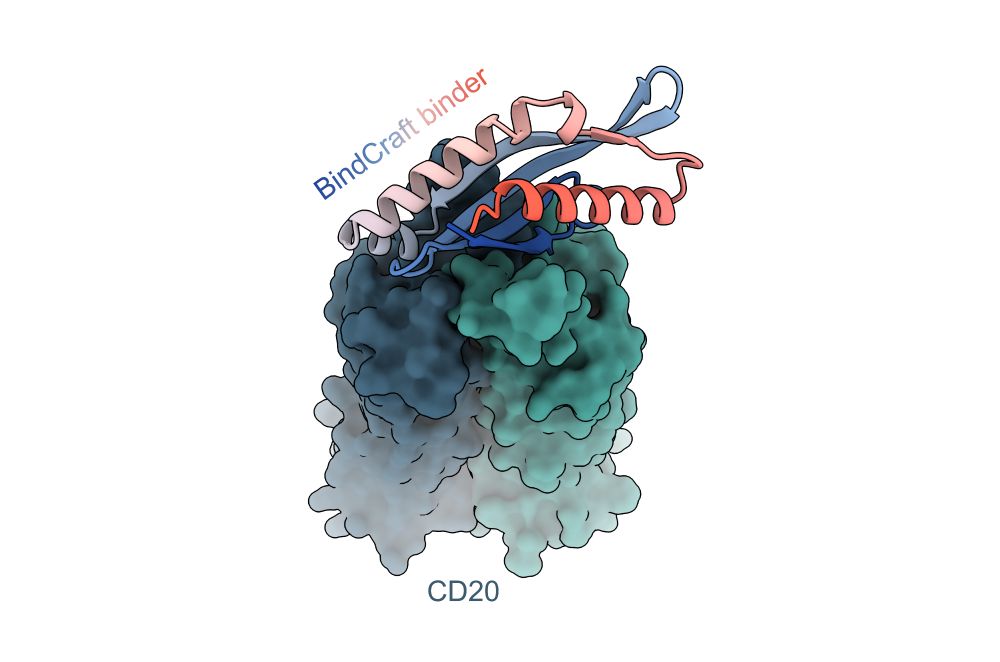
github.com/martinpacesa...
bit.ly/boltz2-pdf

bit.ly/boltz2-pdf






www.biorxiv.org/content/10.1...
P.S.: Not digging the name

www.biorxiv.org/content/10.1...
P.S.: Not digging the name










