www.github.com/LukasHats

www.batcheffect.com/p/the-giant-...

www.batcheffect.com/p/the-giant-...
Happy Halloween! 🎃


Happy Halloween! 🎃
academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social

academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social
www.sciencedirect.com/science/arti...
www.science.org/content/arti...

www.sciencedirect.com/science/arti...
www.science.org/content/arti...

www.nature.com/articles/s4...
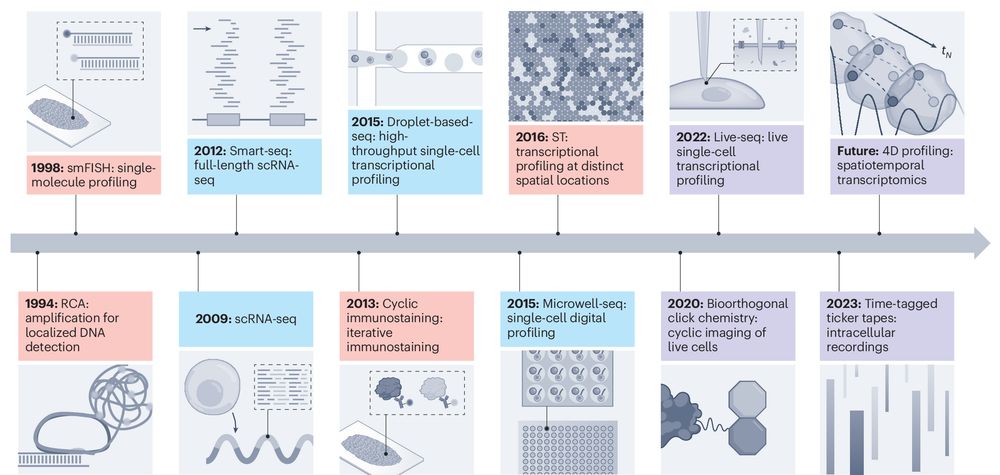
www.nature.com/articles/s4...
Even if it's not a complete and perfect solution, whoever is maintaining the package will help you in get to the right solution and they will be incredibly grateful.
Even if it's not a complete and perfect solution, whoever is maintaining the package will help you in get to the right solution and they will be incredibly grateful.
And congratulations, it's not always easy to jump into an existing codebase and propose changes.
And congratulations, it's not always easy to jump into an existing codebase and propose changes.
This pushed me to completely rewrite the system for generating and plotting boundaries for cell niches.
The new system is now more efficient, consistent, and visually clear.

This pushed me to completely rewrite the system for generating and plotting boundaries for cell niches.
The new system is now more efficient, consistent, and visually clear.
Among some bug fixes, we received our first contribution from an external contributor: @loggas.bsky.social !
He designed a new metric called Relative Component Size (RCS).

Among some bug fixes, we received our first contribution from an external contributor: @loggas.bsky.social !
He designed a new metric called Relative Component Size (RCS).

bmcbioinformatics.biomedcentral.com/articles/10....

bmcbioinformatics.biomedcentral.com/articles/10....
I'm so excited about this!
www.biorxiv.org/content/10.1...

I'm so excited about this!
www.biorxiv.org/content/10.1...
Tree representations for distortion-free visualization and exploratory analysis of single-cell omics data.
The trees are constructed to accurately represent true distances between the objects in the high-dimensional space.
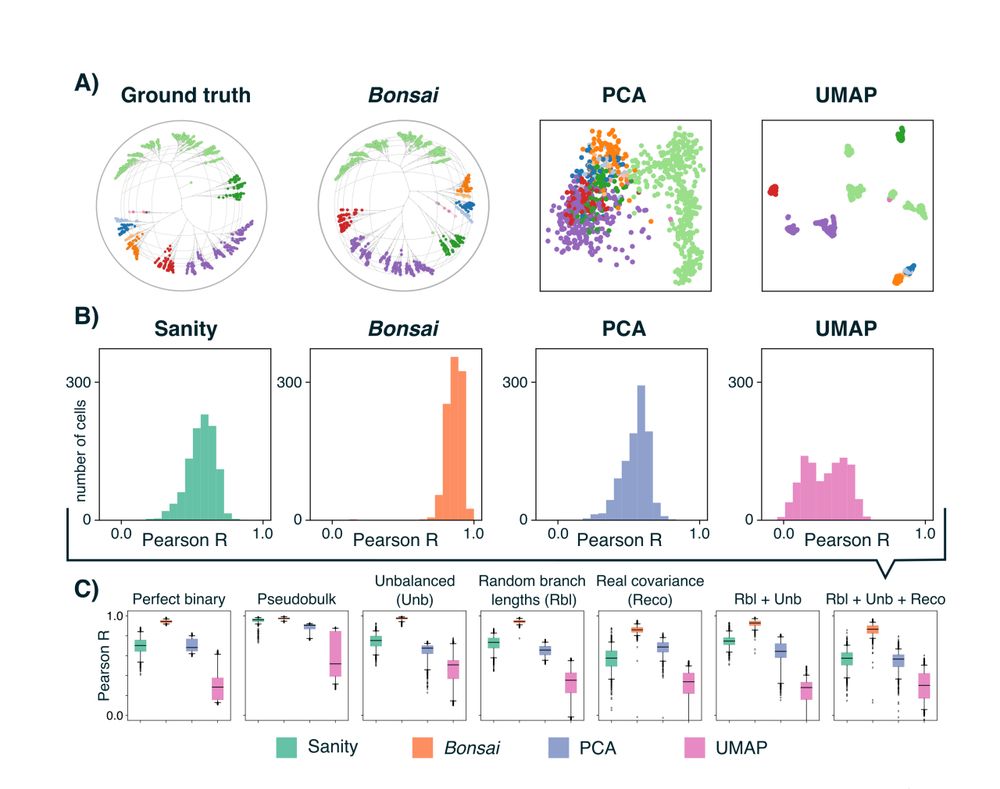
Tree representations for distortion-free visualization and exploratory analysis of single-cell omics data.
The trees are constructed to accurately represent true distances between the objects in the high-dimensional space.
my.corehr.com/pls/uoxrecru...
#academic #jobs #softwaredeveloper #compsci
my.corehr.com/pls/uoxrecru...
#academic #jobs #softwaredeveloper #compsci
🔍 Workflow view – Visualize logical structure of pipelines
⚙️ Process view – See all processes in one place
🌐 Seqera view – Manage connected CEs directly in the IDE
📚 Resources view – Quickly access Copilot, AI tools & training
🔍 Workflow view – Visualize logical structure of pipelines
⚙️ Process view – See all processes in one place
🌐 Seqera view – Manage connected CEs directly in the IDE
📚 Resources view – Quickly access Copilot, AI tools & training
Researchers successfully make miso at the #InternationalSpaceStation, confirming that making fermented foods in space is possible. www.cell.com/iscience/ful...
@medialab.bsky.social Maggie Coblentz
@novo-nordisk.bsky.social Joshua D. Evans
@cp-cell.bsky.social



Researchers successfully make miso at the #InternationalSpaceStation, confirming that making fermented foods in space is possible. www.cell.com/iscience/ful...
@medialab.bsky.social Maggie Coblentz
@novo-nordisk.bsky.social Joshua D. Evans
@cp-cell.bsky.social

Here's my full speech from the Stand Up For Science rally in DC today: www.youtube.com/watch?v=eemj...

Here's my full speech from the Stand Up For Science rally in DC today: www.youtube.com/watch?v=eemj...
Congratulations to @miguelib.bsky.social and Lindsay Caprio for leading this project.
Congratulations to @miguelib.bsky.social and Lindsay Caprio for leading this project.
science.org/toc/sciimmunol/10/105

science.org/toc/sciimmunol/10/105
https://go.nature.com/4bpG2It

https://go.nature.com/4bpG2It


