
Hear Lili Niu at our #HUPO2025 breakfast on how 1152 samples/day and a 200 SPD Evosep Eno + Orbitrap Astral workflow quantified >400 proteins per sample.
Register: www.evosep.com/hupo2025/

Hear Lili Niu at our #HUPO2025 breakfast on how 1152 samples/day and a 200 SPD Evosep Eno + Orbitrap Astral workflow quantified >400 proteins per sample.
Register: www.evosep.com/hupo2025/

#MS #Proteomics #Science @mannlab.bsky.social
#MS #Proteomics #Science @mannlab.bsky.social
www.embopress.org/doi/full/10....

www.embopress.org/doi/full/10....
bsky.app/profile/mann...
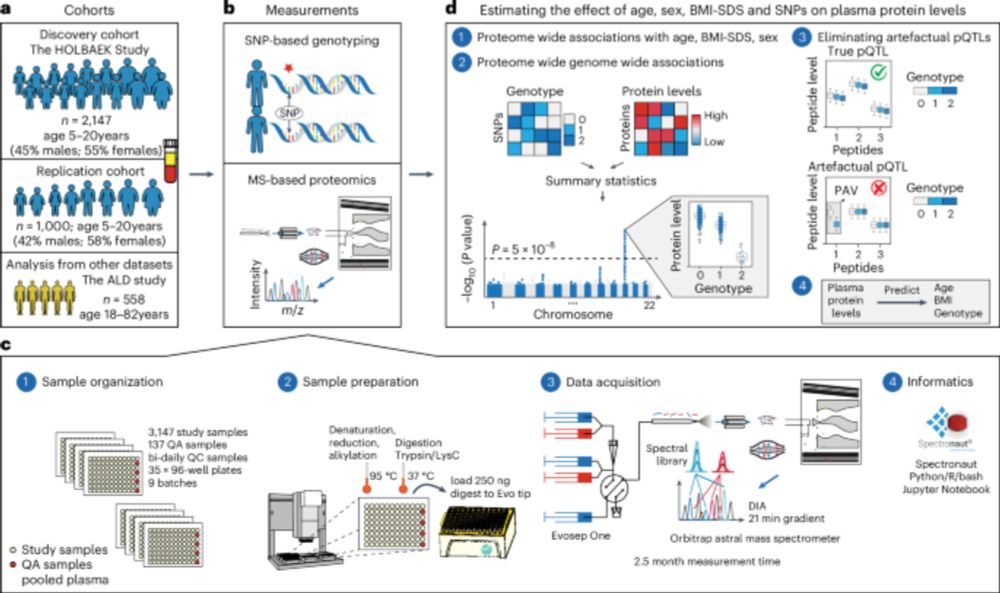
nature.com/articles/s41588-025-02089-2
#PediatricProteomics #pQTL
First author @liliniu.bsky.social explains ⬇️
bsky.app/profile/mann...
nature.com/articles/s41588-025-02089-2
#PediatricProteomics #pQTL
First author @liliniu.bsky.social explains ⬇️

nature.com/articles/s41588-025-02089-2
#PediatricProteomics #pQTL
First author @liliniu.bsky.social explains ⬇️

nature.com/articles/s41588-025-02089-2
#PediatricProteomics #pQTL
First author @liliniu.bsky.social explains ⬇️
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
https://go.nature.com/3WroTb1

https://go.nature.com/3WroTb1


