
Co-Director LSHTM AMR Centre @lshtmamrcentre.bsky.social
holtlab.net | klebnet.org | typhoidgenomics.org | amr.lshtm.ac.uk
Instead I agree with your earlier paper on this topic! www.thelancet.com/journals/lan...

Instead I agree with your earlier paper on this topic! www.thelancet.com/journals/lan...


14 STs caused outbreak clusters in multiple sites, and accounted for 64% of all infections.
These include some of the usual suspects known to cause outbreaks in Europe, US etc including ST307, ST117, ST14, ST15, ST101.

14 STs caused outbreak clusters in multiple sites, and accounted for 64% of all infections.
These include some of the usual suspects known to cause outbreaks in Europe, US etc including ST307, ST117, ST14, ST15, ST101.
The choice of thresholds for 🧬 SNP distance and 📆 temporal distance had little impact on clustering.

The choice of thresholds for 🧬 SNP distance and 📆 temporal distance had little impact on clustering.
Our new preprint shows more than half of #Klebsiella pneumoniae neonatal sepsis cases in African and South Asian are nosocomial, acquired through transmission in neonatal units. #WAAW
doi.org/10.1101/2025...
![figure showing estimated proportion of cases in clusters, for each site. range from 0.04 to 0.93, mean estimate in random effects model is 0.57 [0.46,0.68]](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:q5nky4heg5sfix4get4zh2pw/bafkreibshmuvkuafxwd2uk3r42ss6ma2dhmaidbysgvufnbkbv5cn4guai@jpeg)
Our new preprint shows more than half of #Klebsiella pneumoniae neonatal sepsis cases in African and South Asian are nosocomial, acquired through transmission in neonatal units. #WAAW
doi.org/10.1101/2025...
Most are not on bluesky (yet!)…

Most are not on bluesky (yet!)…


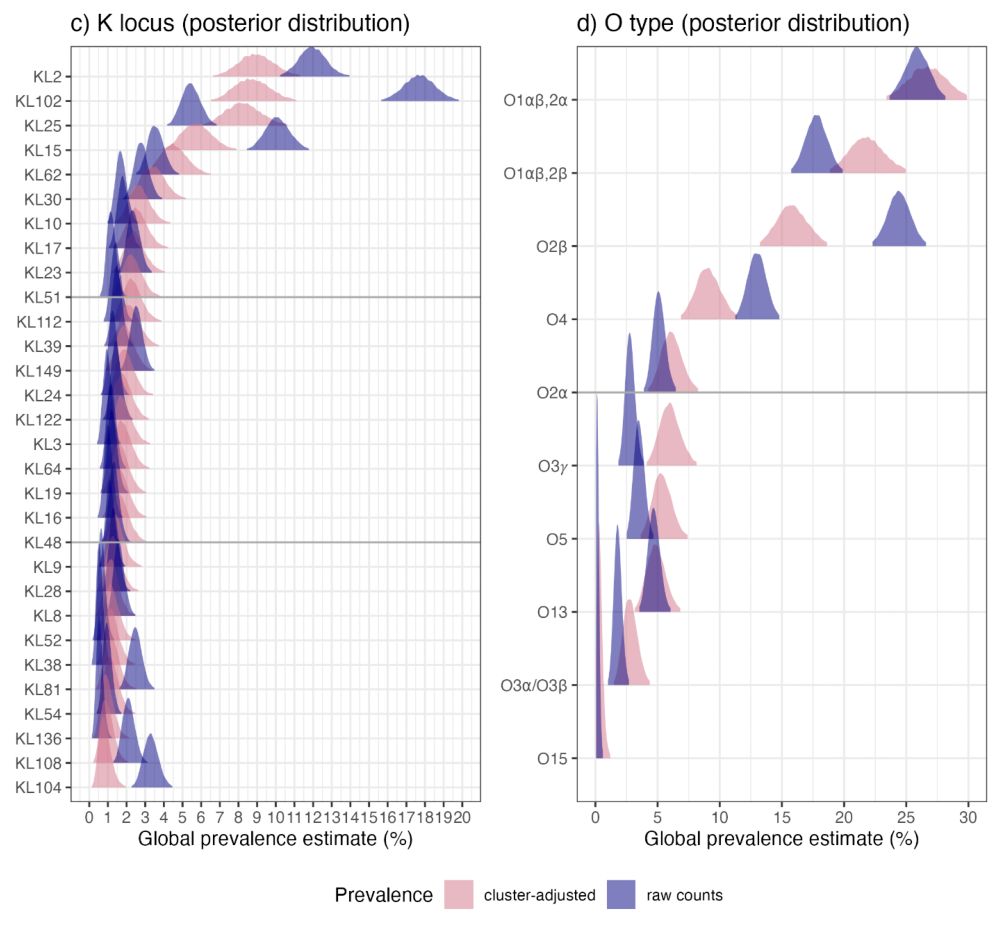
Yes this is a lot of antigens! But it has been done for Streptococcus pneumoniae.

Yes this is a lot of antigens! But it has been done for Streptococcus pneumoniae.



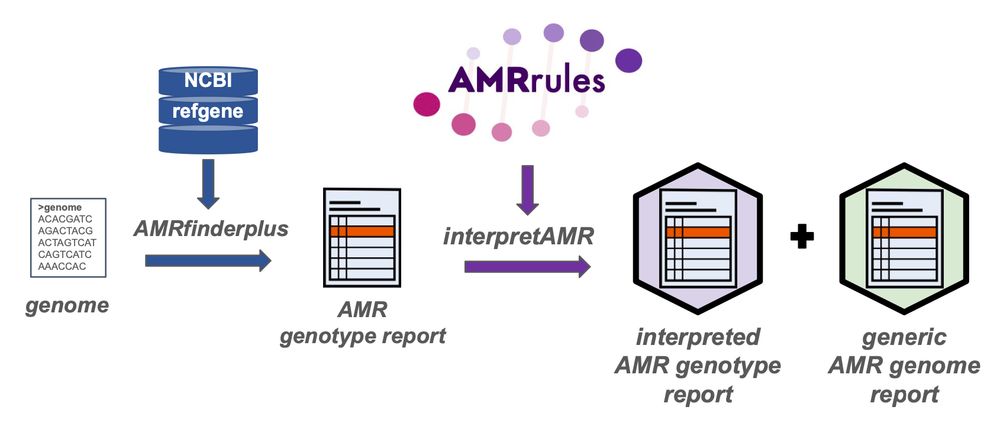
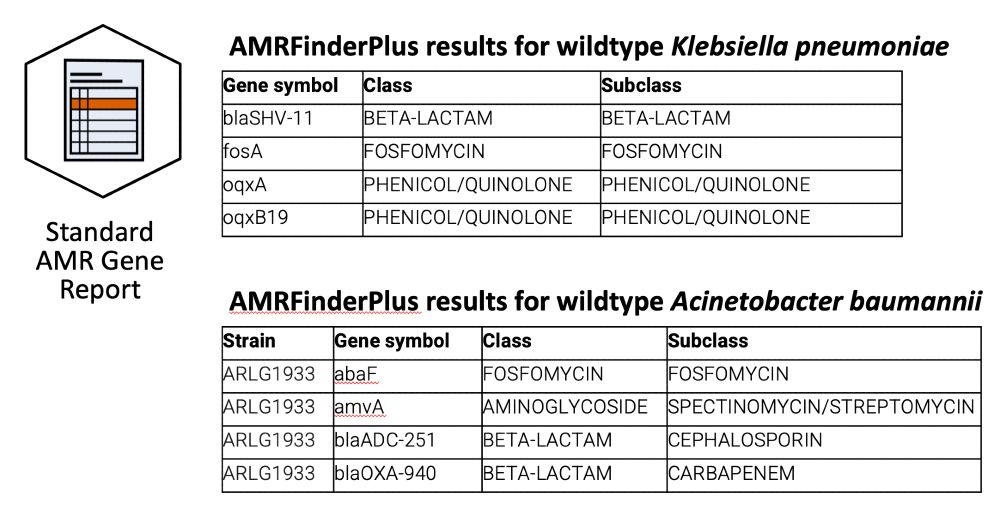
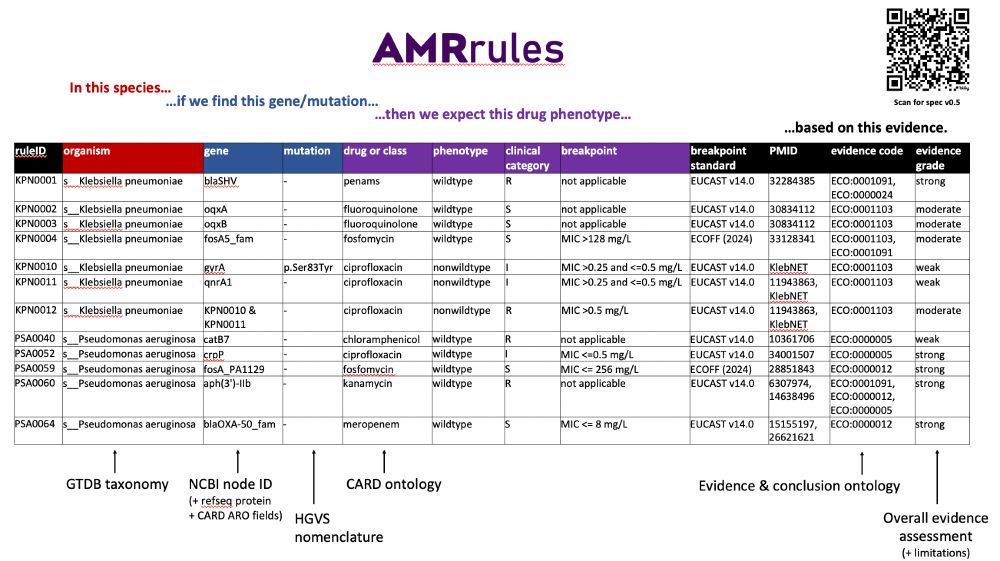
interpretamr.github.io/AMRrules

interpretamr.github.io/AMRrules
#ABPHM25
www.biorxiv.org/content/10.1...

#ABPHM25
www.biorxiv.org/content/10.1...




pubmed.ncbi.nlm.nih.gov/27067320/

pubmed.ncbi.nlm.nih.gov/27067320/
pubmed.ncbi.nlm.nih.gov/26100894/

pubmed.ncbi.nlm.nih.gov/26100894/

