📍Groningen, the Netherlands
Go team!
Go team!
And of course the new parters in crime Eline Smits & @scimarijn.bsky.social, and ever-awesome PI's @irisjonkers.bsky.social & Sebo Withoff.
And of course the new parters in crime Eline Smits & @scimarijn.bsky.social, and ever-awesome PI's @irisjonkers.bsky.social & Sebo Withoff.
With the same iPSC-derived cells, the culture system + signaling regime can shift you from fetal-like to 'adult-like' tissue and from proliferative to highly functional epithelium.
So choosing “organoid vs Transwell vs chip” is not cosmetic – it changes the biology you read out.
With the same iPSC-derived cells, the culture system + signaling regime can shift you from fetal-like to 'adult-like' tissue and from proliferative to highly functional epithelium.
So choosing “organoid vs Transwell vs chip” is not cosmetic – it changes the biology you read out.
👉For drug absorption, metabolism & DDI: you probably want the intestine-on-chip.
👉For developmental biology / early tissue states: organoids are your friend.
👉For certain barrier / ECM / EMT questions: Transwells can be very informative
👉For drug absorption, metabolism & DDI: you probably want the intestine-on-chip.
👉For developmental biology / early tissue states: organoids are your friend.
👉For certain barrier / ECM / EMT questions: Transwells can be very informative
- Organoids cluster closer to fetal intestine
- Intestine-on-chip clusters with pediatric/adult small intestine
- Transwells sit in between but don’t fully capture either extreme
So the chip looks most “adult gut–like” at the transcriptomic level!

- Organoids cluster closer to fetal intestine
- Intestine-on-chip clusters with pediatric/adult small intestine
- Transwells sit in between but don’t fully capture either extreme
So the chip looks most “adult gut–like” at the transcriptomic level!
🔹 Organoids
Strong developmental / fetal signatures
More embryonic patterning pathways
🔹 Transwells
More ECM remodeling, cell cycle, EMT-like features
🔹 Intestine-on-chip
Highest expression of digestion, nutrient transport & drug metabolism genes

🔹 Organoids
Strong developmental / fetal signatures
More embryonic patterning pathways
🔹 Transwells
More ECM remodeling, cell cycle, EMT-like features
🔹 Intestine-on-chip
Highest expression of digestion, nutrient transport & drug metabolism genes
We culture the same iPSC-derived epithelium in EM - DM+D+P as:
- 3D organoids
- 2D Transwell monolayers
- @emulatebio.bsky.social intestine-on-chip under flow
Then we compare their transcriptomes + map them onto in vivo small intestine data.

We culture the same iPSC-derived epithelium in EM - DM+D+P as:
- 3D organoids
- 2D Transwell monolayers
- @emulatebio.bsky.social intestine-on-chip under flow
Then we compare their transcriptomes + map them onto in vivo small intestine data.
The EM–DM + D + P gradient lands perfectly in between:
- Keeps renewal / cell cycle pathways active
- Already shows enhanced digestion and xenobiotic metabolism
So you can balance “crypt-like” proliferation with “villus-like” function by cleverly combining media compositions

The EM–DM + D + P gradient lands perfectly in between:
- Keeps renewal / cell cycle pathways active
- Already shows enhanced digestion and xenobiotic metabolism
So you can balance “crypt-like” proliferation with “villus-like” function by cleverly combining media compositions
First organoids:
Compared to EM, the DM + D + P massively boosts genes for:
- Nutrient digestion
- Transporters
- Drug metabolism
These iPSC-derived cells can become strongly enterocyte-like!

First organoids:
Compared to EM, the DM + D + P massively boosts genes for:
- Nutrient digestion
- Transporters
- Drug metabolism
These iPSC-derived cells can become strongly enterocyte-like!
On Transwells we installed a growth factor gradient for this:
Basolateral: EM
Apical: DM + D _or_ DM + D + P
This indeed keeps a proliferative compartment and generates differentiated cells with tight junctions and good barrier properties.

On Transwells we installed a growth factor gradient for this:
Basolateral: EM
Apical: DM + D _or_ DM + D + P
This indeed keeps a proliferative compartment and generates differentiated cells with tight junctions and good barrier properties.
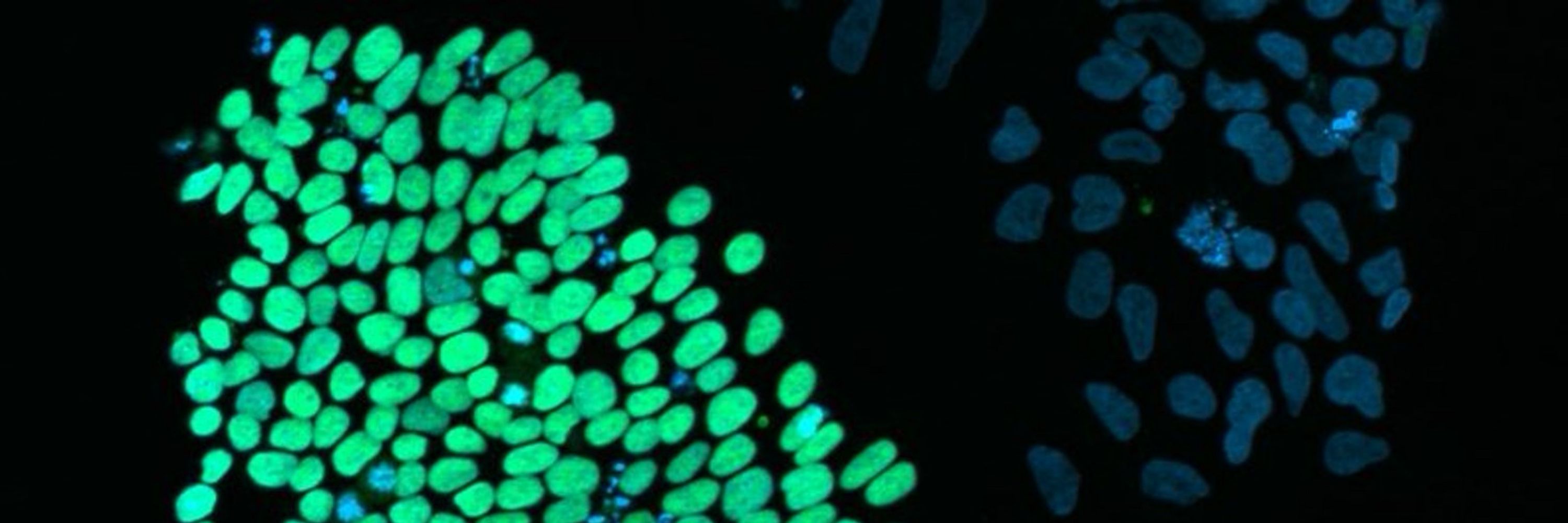
- EM → lots of MKI67⁺ proliferative cells
- DM / DM + D → more enterocytes + goblet cells
- DM + D + P → differentiated epithelium with detectable enteroendocrine cells
(Paneth(-like) cells behave differently depending on the platform)

- EM → lots of MKI67⁺ proliferative cells
- DM / DM + D → more enterocytes + goblet cells
- DM + D + P → differentiated epithelium with detectable enteroendocrine cells
(Paneth(-like) cells behave differently depending on the platform)
- EM = expansion (high WNT, BMP inhibition) → stem/transit-amplifying
- DM = differentiation
- DM + D = + NOTCH inhibitor → more secretory cells
- DM + D + P = + MEK inhibitor → more enteroendocrine cells
- EM = expansion (high WNT, BMP inhibition) → stem/transit-amplifying
- DM = differentiation
- DM + D = + NOTCH inhibitor → more secretory cells
- DM + D + P = + MEK inhibitor → more enteroendocrine cells
This lets us push cells from a proliferative, stem-like state toward mature epithelial lineages.
This lets us push cells from a proliferative, stem-like state toward mature epithelial lineages.
But the same starting cells can behave very differently depending on whether you use them as 3D organoids, 2D monolayers on Transwells or on an intestine-on-chip.
So we asked: how big is that difference, really?
www.cell.com/stem-cell-re...

But the same starting cells can behave very differently depending on whether you use them as 3D organoids, 2D monolayers on Transwells or on an intestine-on-chip.
So we asked: how big is that difference, really?
www.cell.com/stem-cell-re...
But cant LLMs here help kickstart the decoupling of journals and PR to maintain the quality check of the current Status Quo of PR. and once that has happened, we can move to this post publication ongoing PR sytem
But cant LLMs here help kickstart the decoupling of journals and PR to maintain the quality check of the current Status Quo of PR. and once that has happened, we can move to this post publication ongoing PR sytem

