Joy Fan
@jlfan.bsky.social
PhD candidate in BME @Columbia | computational #cancer research at http://azizilab.com
#Echidna is now publicly available at github.com/azizilab/ech..., and may also be installed on PyPI and Bioconda! We hope you will join us in understanding the role of CNAs in phenotypic plasticity!
GitHub - azizilab/echidna: Mapping genotype to phenotype through joint probabilistic modeling of single-cell gene expression and chromosomal copy number variation
Mapping genotype to phenotype through joint probabilistic modeling of single-cell gene expression and chromosomal copy number variation - azizilab/echidna
github.com
December 18, 2024 at 1:37 PM
#Echidna is now publicly available at github.com/azizilab/ech..., and may also be installed on PyPI and Bioconda! We hope you will join us in understanding the role of CNAs in phenotypic plasticity!
📢 TL;DR: #Echidna 🦔integrates scRNA-seq & WGS to decouple gene dosage effects on phenotypic plasticity, tracks clonal evolution, and identifies resistance drivers in tumors.
Read more here: www.biorxiv.org/content/10.1...
Let’s unravel tumor evolution, one clone at a time! 7/7
Read more here: www.biorxiv.org/content/10.1...
Let’s unravel tumor evolution, one clone at a time! 7/7
www.biorxiv.org
December 18, 2024 at 1:31 PM
📢 TL;DR: #Echidna 🦔integrates scRNA-seq & WGS to decouple gene dosage effects on phenotypic plasticity, tracks clonal evolution, and identifies resistance drivers in tumors.
Read more here: www.biorxiv.org/content/10.1...
Let’s unravel tumor evolution, one clone at a time! 7/7
Read more here: www.biorxiv.org/content/10.1...
Let’s unravel tumor evolution, one clone at a time! 7/7
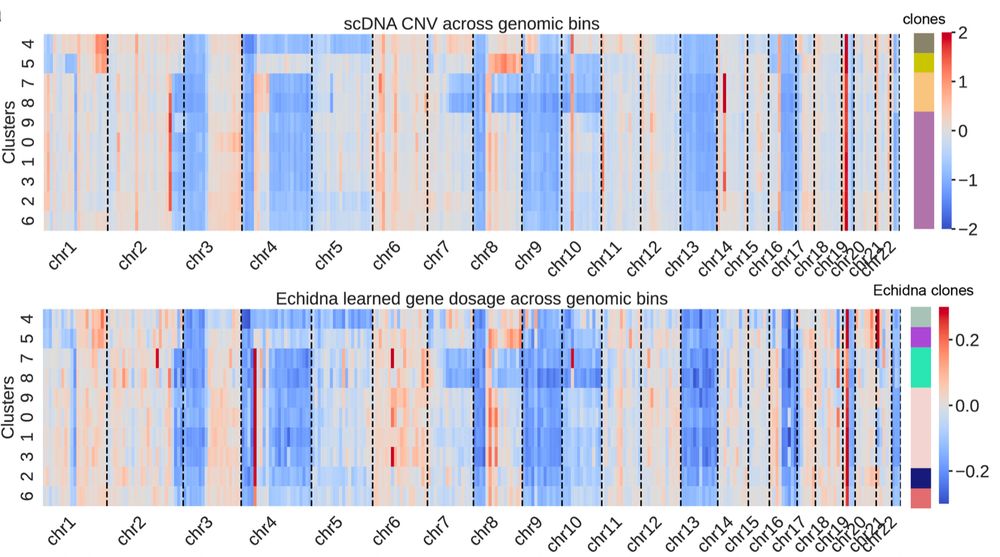
🌟 Why is this impactful? #Echidna doesn’t require joint scDNA/RNA sequencing (rare/expensive). Instead, it pairs scRNA-seq & WGS—readily obtainable from clinical samples. It’s scalable & can reveal mechanisms of cancer progression & treatment response! 🧬 6/
December 18, 2024 at 1:31 PM
🌟 Why is this impactful? #Echidna doesn’t require joint scDNA/RNA sequencing (rare/expensive). Instead, it pairs scRNA-seq & WGS—readily obtainable from clinical samples. It’s scalable & can reveal mechanisms of cancer progression & treatment response! 🧬 6/
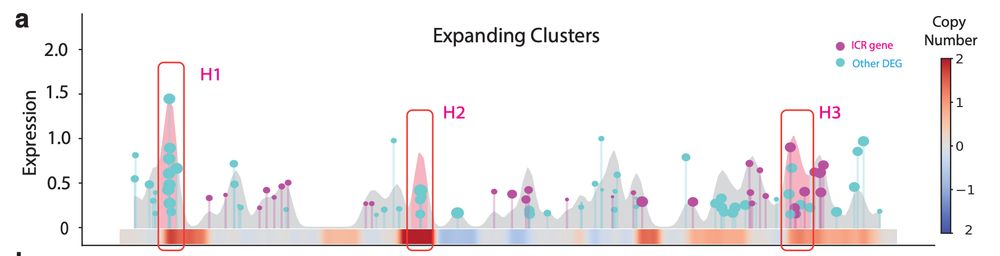
A Case Study in Melanoma: 🧬 Clones resistant to anti-PD-1 therapy showed clusters of phenotype-defining genes in hotspots of amplification! Including:
S100 family and MHC-II genes. #Echidna also disentangles intrinsic (CNA-driven) vs extrinsic expression immune signaling!
S100 family and MHC-II genes. #Echidna also disentangles intrinsic (CNA-driven) vs extrinsic expression immune signaling!
December 18, 2024 at 1:31 PM
A Case Study in Melanoma: 🧬 Clones resistant to anti-PD-1 therapy showed clusters of phenotype-defining genes in hotspots of amplification! Including:
S100 family and MHC-II genes. #Echidna also disentangles intrinsic (CNA-driven) vs extrinsic expression immune signaling!
S100 family and MHC-II genes. #Echidna also disentangles intrinsic (CNA-driven) vs extrinsic expression immune signaling!
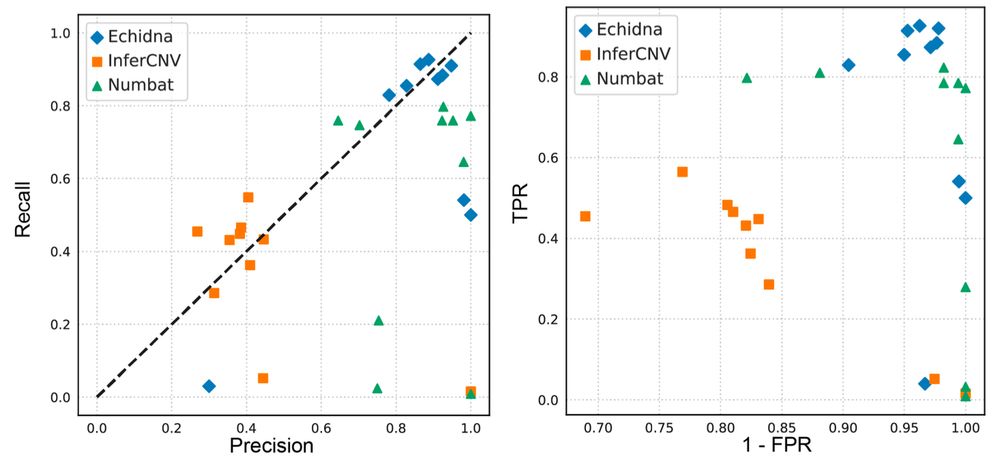
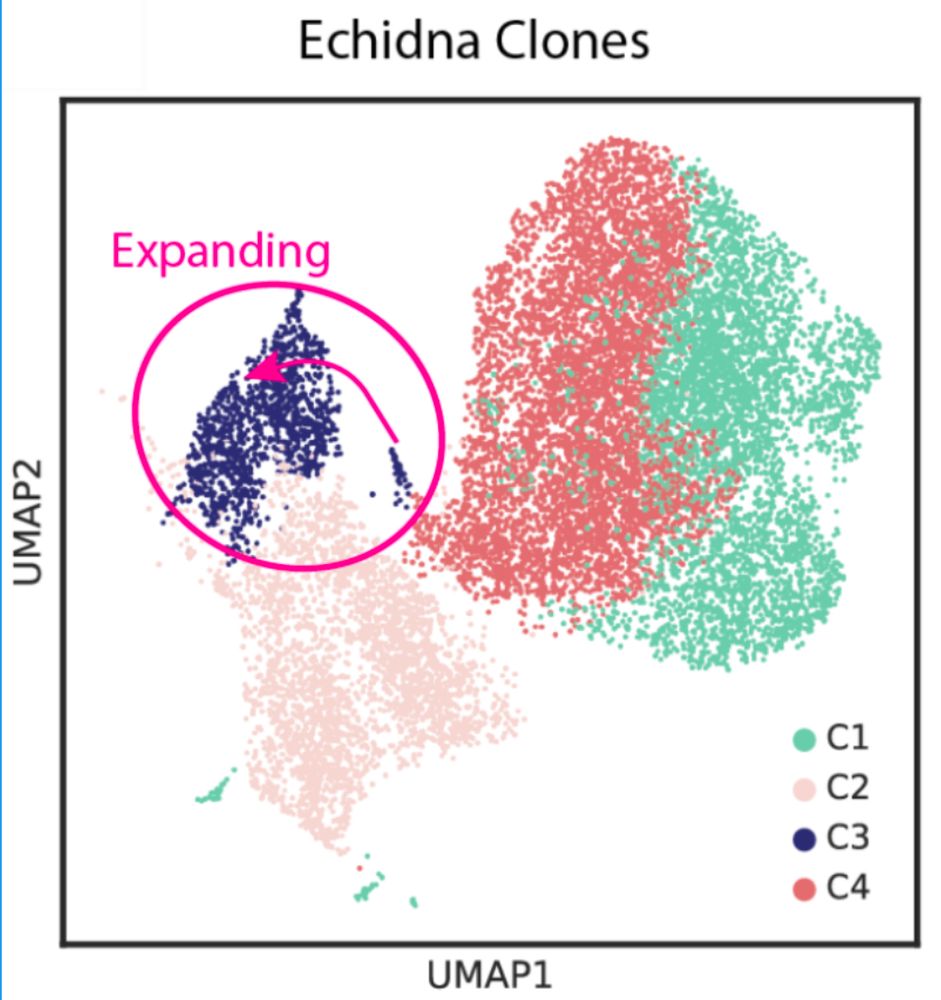
What did we find? Applied to tumor samples: 1️⃣ Clonal structure reconstructed with higher accuracy vs InferCNV/Numbat. 2️⃣ Temporal dynamics captured: tracks clonal evolution pre/post therapy. 3️⃣ Drivers of resistance identified using GDX in genomic hotspots. 4/
December 18, 2024 at 1:31 PM
What did we find? Applied to tumor samples: 1️⃣ Clonal structure reconstructed with higher accuracy vs InferCNV/Numbat. 2️⃣ Temporal dynamics captured: tracks clonal evolution pre/post therapy. 3️⃣ Drivers of resistance identified using GDX in genomic hotspots. 4/
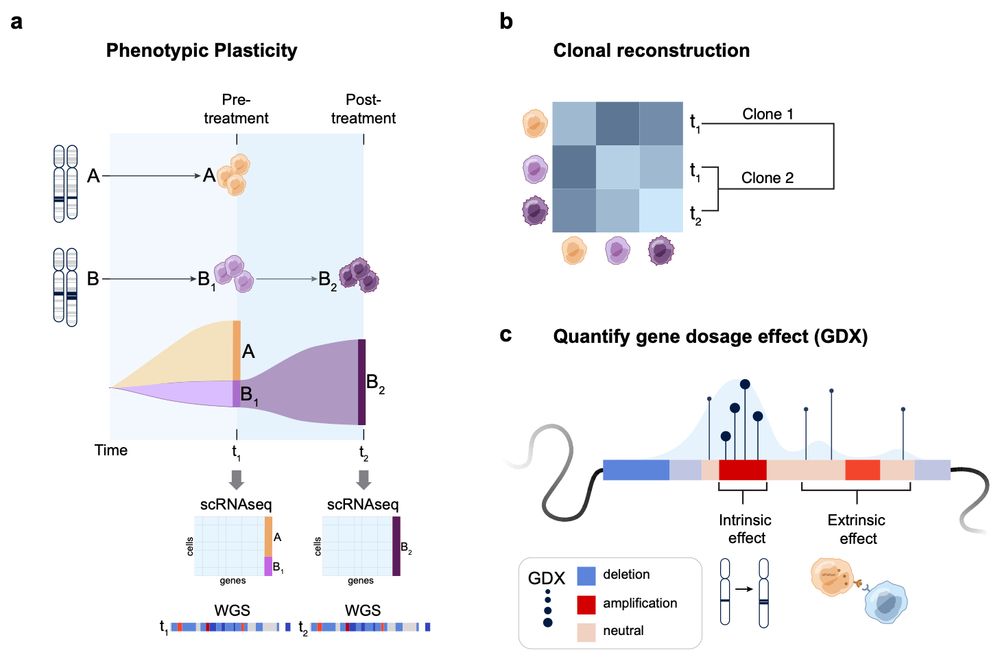
How it works: #Echidna models single-cell RNA & bulk WGS data using a Bayesian hierarchical framework. Key features:
Deconvolves CNA profiles
Tracks clones & their phenotypic states over time
Introduces GDX: a metric quantifying gene dosage effects! 3/
Deconvolves CNA profiles
Tracks clones & their phenotypic states over time
Introduces GDX: a metric quantifying gene dosage effects! 3/
December 18, 2024 at 1:31 PM
How it works: #Echidna models single-cell RNA & bulk WGS data using a Bayesian hierarchical framework. Key features:
Deconvolves CNA profiles
Tracks clones & their phenotypic states over time
Introduces GDX: a metric quantifying gene dosage effects! 3/
Deconvolves CNA profiles
Tracks clones & their phenotypic states over time
Introduces GDX: a metric quantifying gene dosage effects! 3/
📌 Why Echidna? Phenotypic plasticity (cell adaptability) is critical in cancer progression & resistance. But how much of this plasticity is driven by gene dosage vs external cues?
Echidna uncouples these effects, bridging genome & transcriptome across timepoints!
Echidna uncouples these effects, bridging genome & transcriptome across timepoints!
December 18, 2024 at 1:31 PM
📌 Why Echidna? Phenotypic plasticity (cell adaptability) is critical in cancer progression & resistance. But how much of this plasticity is driven by gene dosage vs external cues?
Echidna uncouples these effects, bridging genome & transcriptome across timepoints!
Echidna uncouples these effects, bridging genome & transcriptome across timepoints!

