I like thinking about computational biological sequence analysis and its applications to metagenomics.
https://jim-shaw-bluenote.github.io
So much to explore... for example, we compared 6 circular TM7 bacteria of > 93% ANI assembled from a single oral metagenome.
10 / N

So much to explore... for example, we compared 6 circular TM7 bacteria of > 93% ANI assembled from a single oral metagenome.
10 / N
1. The distinct ermFs are spreading on two distinct MGEs.
2. There is even strain specificity, only 1/6 P. copri had it!
9 / N

1. The distinct ermFs are spreading on two distinct MGEs.
2. There is even strain specificity, only 1/6 P. copri had it!
9 / N
For example, mobile genetic elements, AMR genes that are hard to bin and assemble with short reads...?
8/N

For example, mobile genetic elements, AMR genes that are hard to bin and assemble with short reads...?
8/N
It turns out myloasm can recover more near-complete contigs than other ONT methods.
For a gut sample, it could assemble _6 different Prevotella copri genomes_ into single contigs, whereas other methods struggled.
7 / N

It turns out myloasm can recover more near-complete contigs than other ONT methods.
For a gut sample, it could assemble _6 different Prevotella copri genomes_ into single contigs, whereas other methods struggled.
7 / N
For jointly-sequenced gut samples (thanks to public data from @jjminich.bsky.social), ONT can assemble _more_ circular contigs than HiFi.
This is thanks to ~3-5x increases in circular contigs relative to previous methods.
5 / N

For jointly-sequenced gut samples (thanks to public data from @jjminich.bsky.social), ONT can assemble _more_ circular contigs than HiFi.
This is thanks to ~3-5x increases in circular contigs relative to previous methods.
5 / N
For ONT R10.4 data, myloasm stands out, especially for retrieving circular, complete contigs.
For HiFi, metaMDBG is very competitive. hifiasm-meta is still strong for gut samples. 4 / N

For ONT R10.4 data, myloasm stands out, especially for retrieving circular, complete contigs.
For HiFi, metaMDBG is very competitive. hifiasm-meta is still strong for gut samples. 4 / N
Anyways, we show this leads to good assemblies, especially when similar genomes (within-species diversity) are present. See results for mock ONT R10.4 synthetic community:
3 / N

Anyways, we show this leads to good assemblies, especially when similar genomes (within-species diversity) are present. See results for mock ONT R10.4 synthetic community:
3 / N
We often use SNPs to disentangle similar genomic regions ("inexact repeats", e.g. haplotypes).
But in assembly, no reference exists, so we use a context-free k-mer representation instead ("SNPmers").
2 / N

We often use SNPs to disentangle similar genomic regions ("inexact repeats", e.g. haplotypes).
But in assembly, no reference exists, so we use a context-free k-mer representation instead ("SNPmers").
2 / N
We try to be upfront about this and discuss it here myloasm-docs.github.io/qc/. We provide additional info for plotting and curation too.
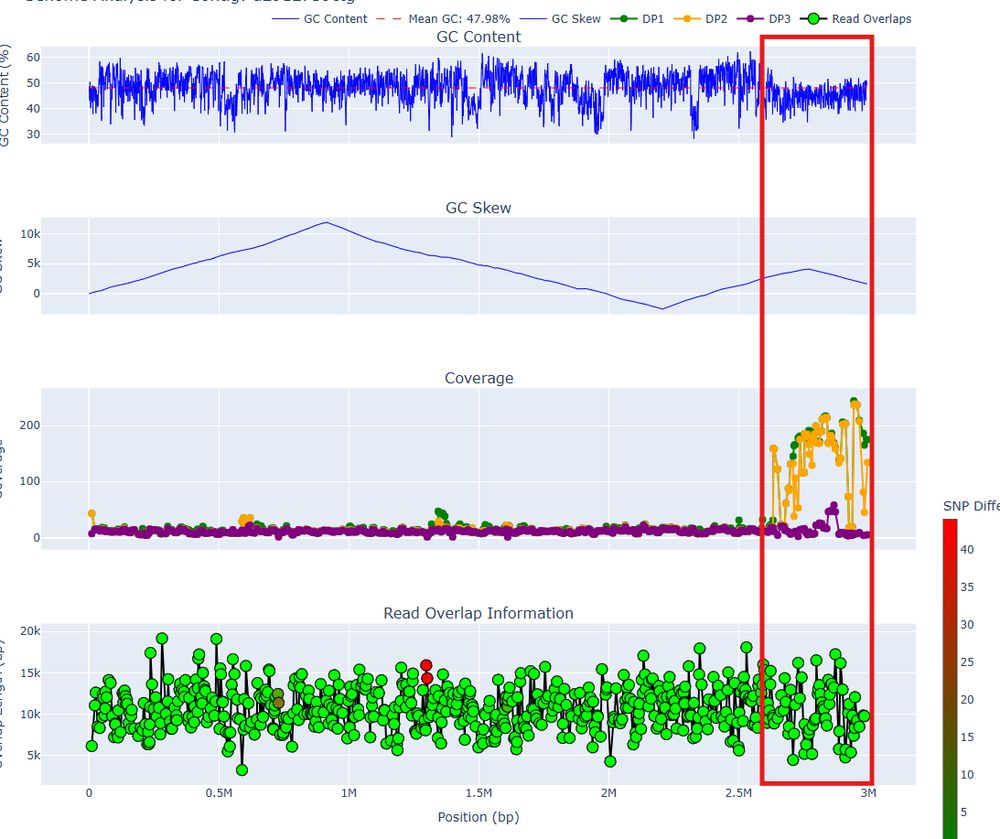
We try to be upfront about this and discuss it here myloasm-docs.github.io/qc/. We provide additional info for plotting and curation too.
We assembled 6 single-contig Prevotella copri genomes of > 97% ANI for one metagenome. 4 of them were circular.
(The largest metaFlye P. copri contig was 13.4% complete)
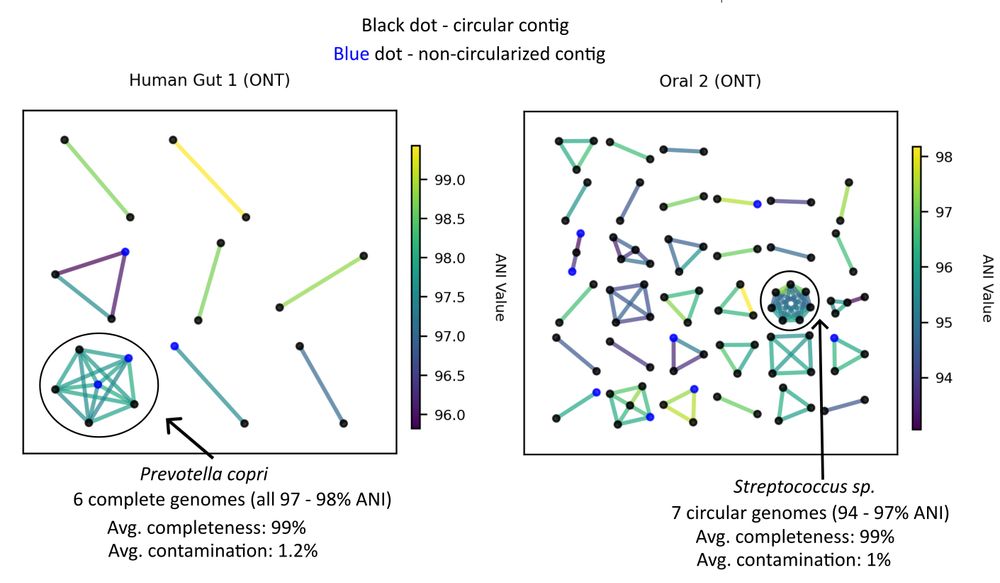
We assembled 6 single-contig Prevotella copri genomes of > 97% ANI for one metagenome. 4 of them were circular.
(The largest metaFlye P. copri contig was 13.4% complete)
The main strength of myloasm: it seems like it can assemble more circularized complete genomes than before, and on diverse metagenomes.

The main strength of myloasm: it seems like it can assemble more circularized complete genomes than before, and on diverse metagenomes.
We applied devider to a bovine gut metagenome enriched for AMR... and we found lots of interesting mosaic haplotypes. See the recombination blocks in the image (found by GARD).
4/5

We applied devider to a bovine gut metagenome enriched for AMR... and we found lots of interesting mosaic haplotypes. See the recombination blocks in the image (found by GARD).
4/5
Technique inspired by a great paper from Zhou et al. for diploid haplotyping (www.nature.com/articles/s41...)
3/5
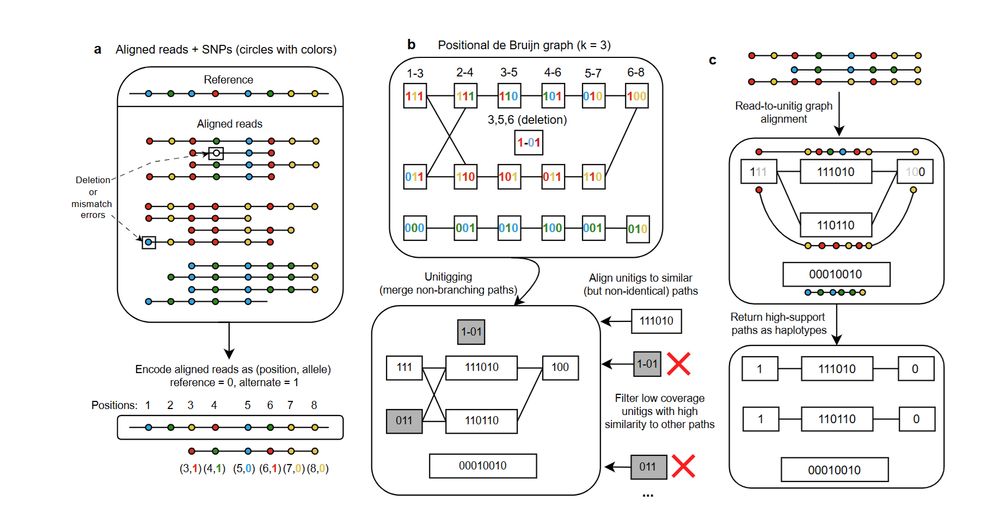
Technique inspired by a great paper from Zhou et al. for diploid haplotyping (www.nature.com/articles/s41...)
3/5
Main points:
(1) Works best with sequence length ~= read length
(2) Does not require prior knowledge of # of haplotypes (can be many!)
(3) Extremely fast (> 20,000x coverage is ok!)
(4) Requires reference
2/5

Main points:
(1) Works best with sequence length ~= read length
(2) Does not require prior knowledge of # of haplotypes (can be many!)
(3) Extremely fast (> 20,000x coverage is ok!)
(4) Requires reference
2/5
Sensitivity for low-abund species is much improved for Illumina now (thanks @fplazaonate for showing issue). bioRxiv updated with a few new benchmarks too.

Sensitivity for low-abund species is much improved for Illumina now (thanks @fplazaonate for showing issue). bioRxiv updated with a few new benchmarks too.

