London · briscoelab.org

More ventral markers (Nkx2.2) appear later but domain sizes remain constant
This suggests progressive fate assignment reflects gene regulatory dynamics, not gradient dynamics
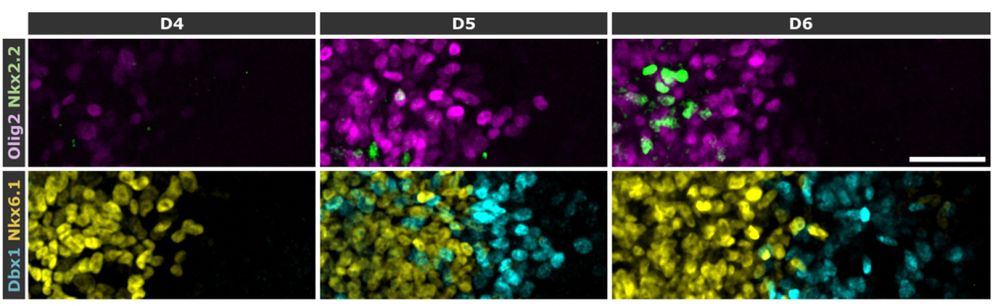
More ventral markers (Nkx2.2) appear later but domain sizes remain constant
This suggests progressive fate assignment reflects gene regulatory dynamics, not gradient dynamics
The system provides a quantitative readout for morphogen modulator activity

The system provides a quantitative readout for morphogen modulator activity
Membrane-tethered Shh-CD4 abolished long-range signalling
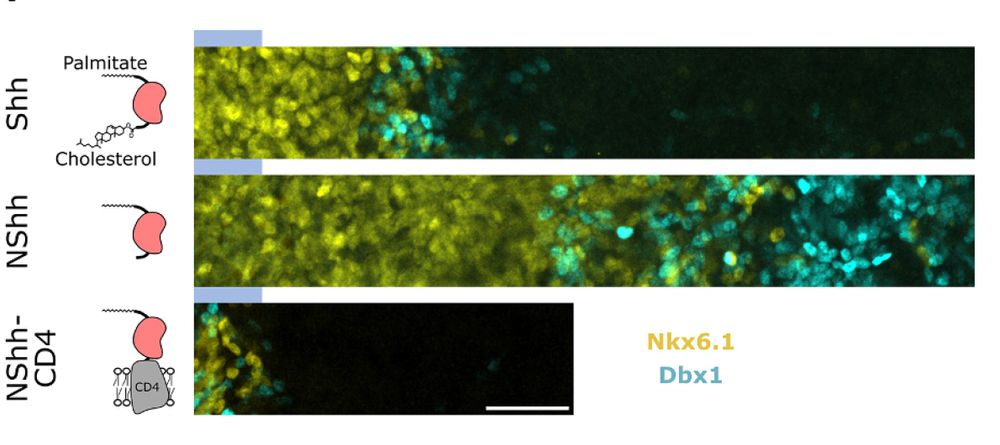
Membrane-tethered Shh-CD4 abolished long-range signalling
Suggests Shh's effective diffusion rate: ~0.125 μm²/s, remarkably similar to Dpp/Wingless in Drosophila
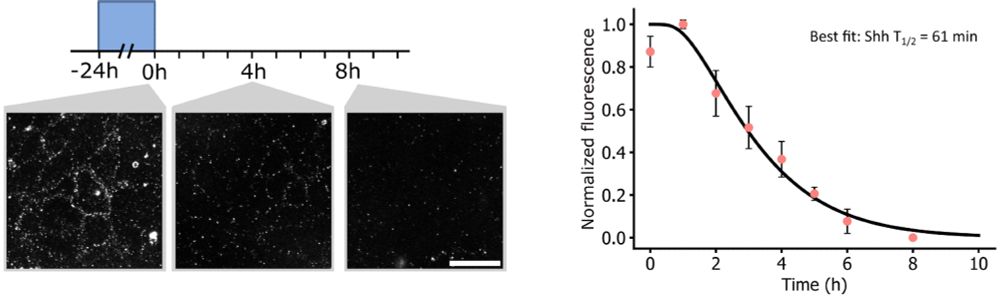
Suggests Shh's effective diffusion rate: ~0.125 μm²/s, remarkably similar to Dpp/Wingless in Drosophila
Spatially restricted light illumination recreated long-range Shh gradients that faithfully recapitulated neural tube patterning

Spatially restricted light illumination recreated long-range Shh gradients that faithfully recapitulated neural tube patterning
By titrating Shld-1 concentration, we can precisely tune activation levels - something challenging to achieve with light alone
It's a dimmer switch, not just a toggle

By titrating Shld-1 concentration, we can precisely tune activation levels - something challenging to achieve with light alone
It's a dimmer switch, not just a toggle
To get around this we fused the light-responsive transcription factor GAVPO to a small-molecule responsive degron domain (DD-GAVPO)
Best of both worlds: chemo-optogenetics
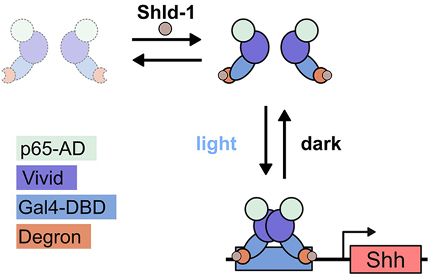
To get around this we fused the light-responsive transcription factor GAVPO to a small-molecule responsive degron domain (DD-GAVPO)
Best of both worlds: chemo-optogenetics
The mathematical structure underlying cell fate decisions appear to be fundamental features of vertebrate neural tube development

The mathematical structure underlying cell fate decisions appear to be fundamental features of vertebrate neural tube development
When we tested new patterns of Sonic Hedgehog signalling experimentally, the predictions matched xpts
This is true predictive biology, not just curve fitting
When we tested new patterns of Sonic Hedgehog signalling experimentally, the predictions matched xpts
This is true predictive biology, not just curve fitting
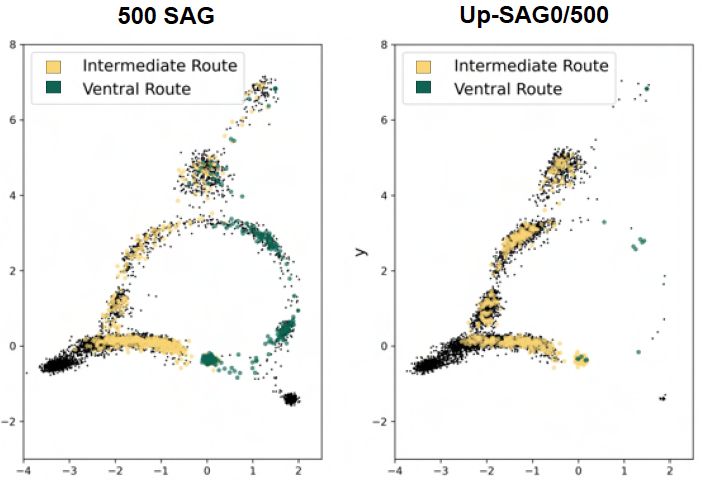
Cells that initially diverge into different lineages can converge through multiple routes to the same fate
This challenges the traditional "branching tree" view of development
Cells that initially diverge into different lineages can converge through multiple routes to the same fate
This challenges the traditional "branching tree" view of development
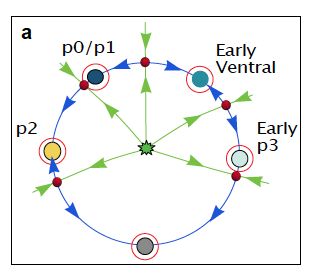
• Direct transitions (fold bifurcations)
• Binary flip decisions
• Binary choice decisions
These mathematical structures explain how signalling controls cell fate allocation
• Direct transitions (fold bifurcations)
• Binary flip decisions
• Binary choice decisions
These mathematical structures explain how signalling controls cell fate allocation
It bridges microscopic gene expression to macroscopic cell behaviour

It bridges microscopic gene expression to macroscopic cell behaviour
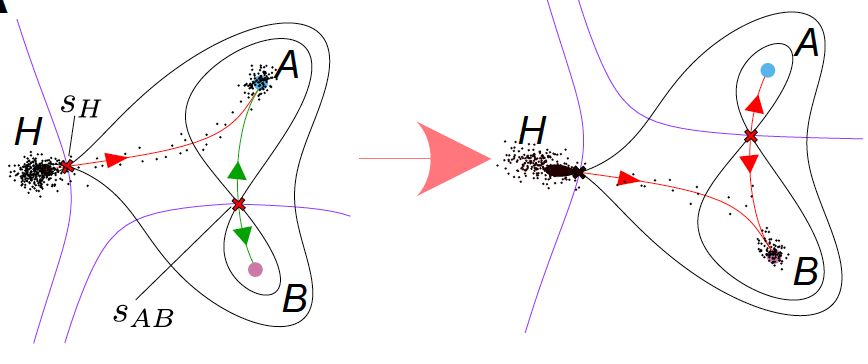
• Identifies stable cell states as "attractor clusters"
• Maps transition pathways between states
• Builds predictive mathematical models

• Identifies stable cell states as "attractor clusters"
• Maps transition pathways between states
• Builds predictive mathematical models
Most approaches describe what we see, not how it happens
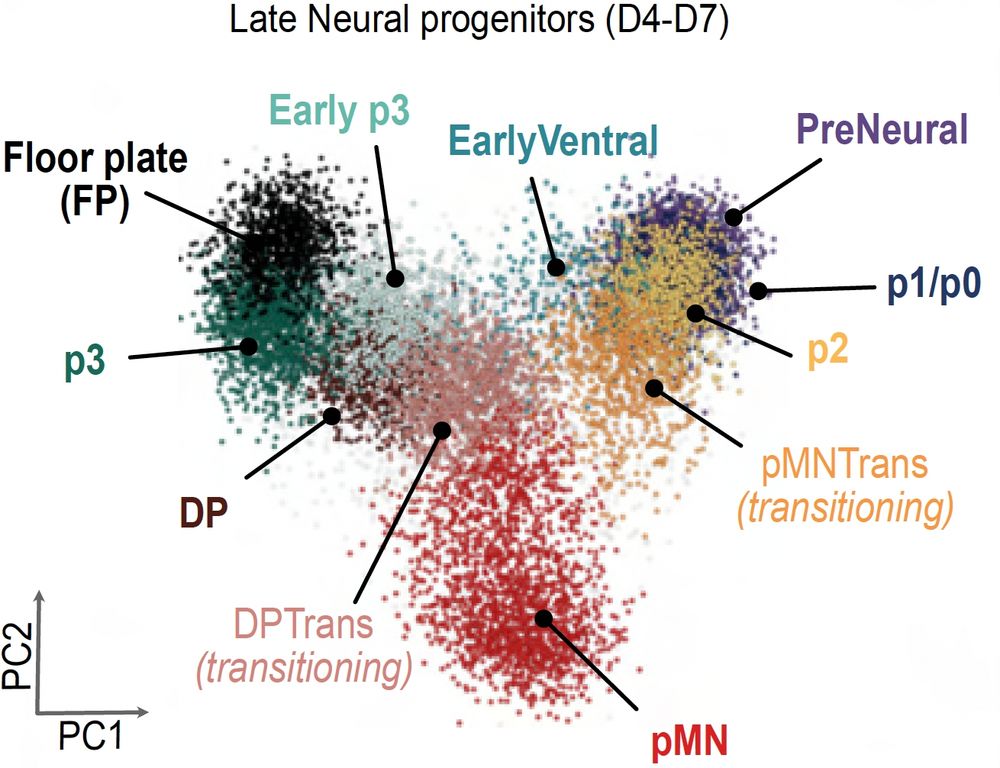
Most approaches describe what we see, not how it happens
Looking forward to a few days at #GERN2025
www.mbl.edu/education/ad...
Looking forward to a few days at #GERN2025
www.mbl.edu/education/ad...

The wonderful Vanessa Ribes discussing recent work on txn regulation by Pax3/7 in neural progenitors #biologists100
www.biorxiv.org/content/10.1...

The wonderful Vanessa Ribes discussing recent work on txn regulation by Pax3/7 in neural progenitors #biologists100
www.biorxiv.org/content/10.1...
And reveals how transcriptional elongation control via MLLT3/SEC helps coordinate rapid cell fate transitions during development
And reveals how transcriptional elongation control via MLLT3/SEC helps coordinate rapid cell fate transitions during development
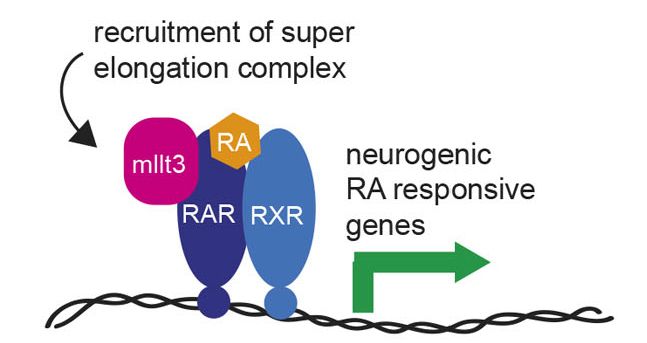
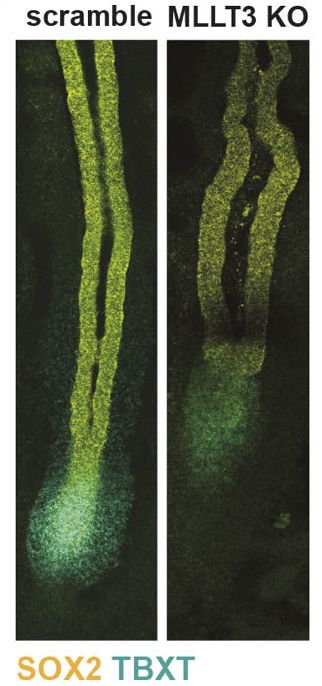
This positions MLLT3 as a link between NMP maintenance and neural differentiation
This positions MLLT3 as a link between NMP maintenance and neural differentiation
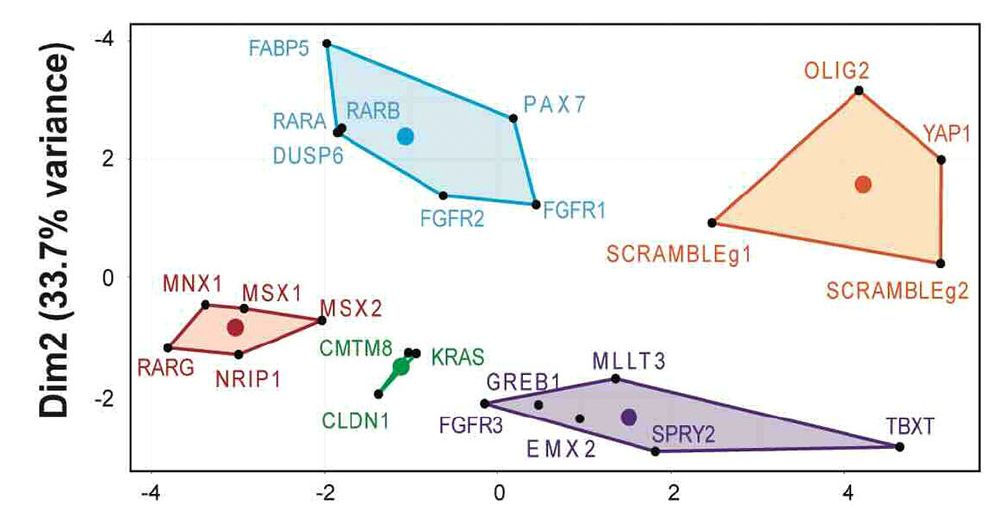
Suggests MLLT3 plays a previously unknown role in neural specification
Suggests MLLT3 plays a previously unknown role in neural specification
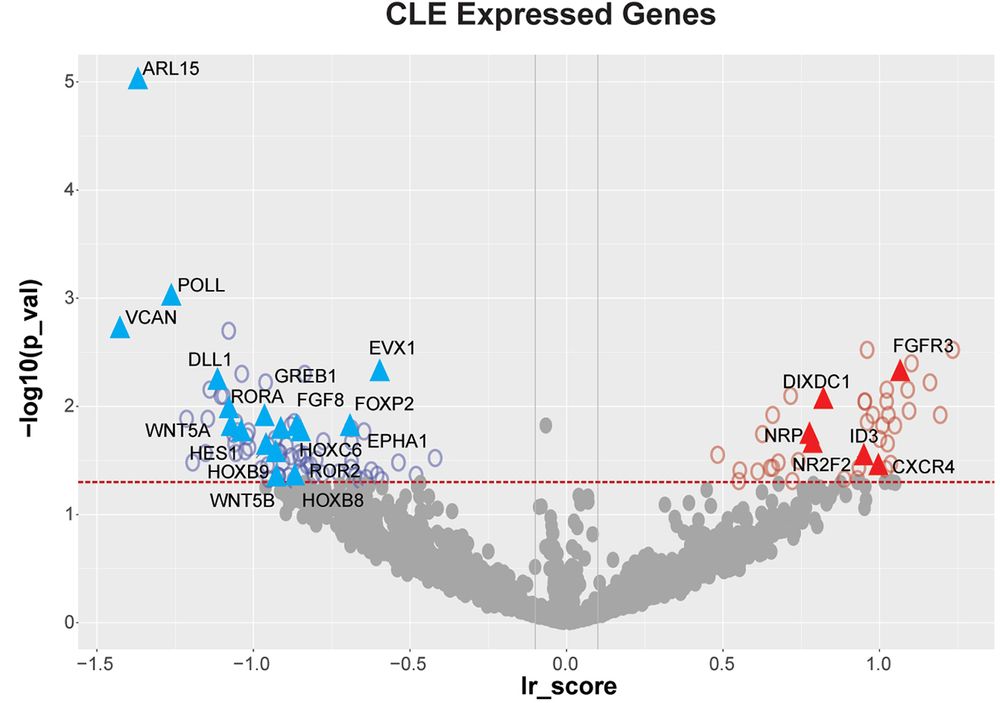
FGF and RAR knockouts accumulated cells in CLE, consistent with their roles in differentiation
FGF and RAR knockouts accumulated cells in CLE, consistent with their roles in differentiation
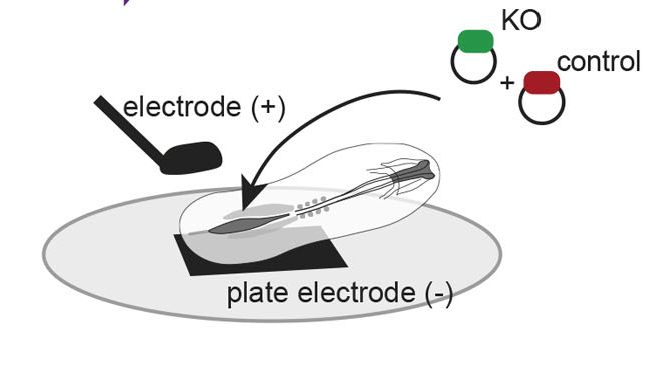
Then performed a screen targeting 25 genes with 4 guides each (102 total guides including controls) in the CLE
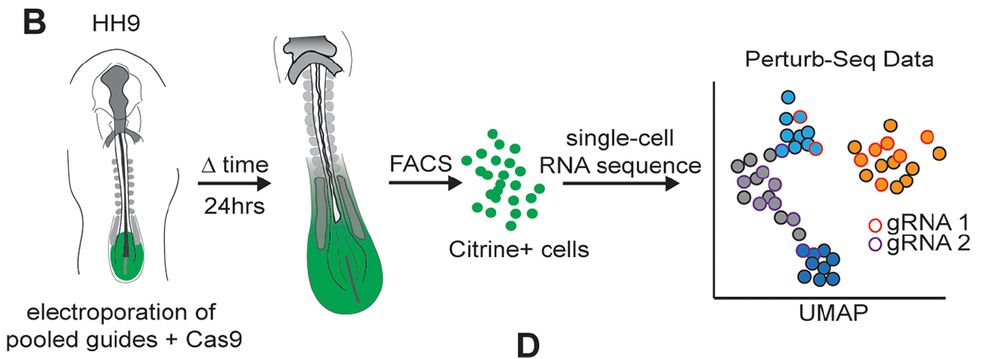
Then performed a screen targeting 25 genes with 4 guides each (102 total guides including controls) in the CLE
Key refinement: incorporating capture sequences into gRNAs to enable single-cell RNA-seq tracking of perturbations
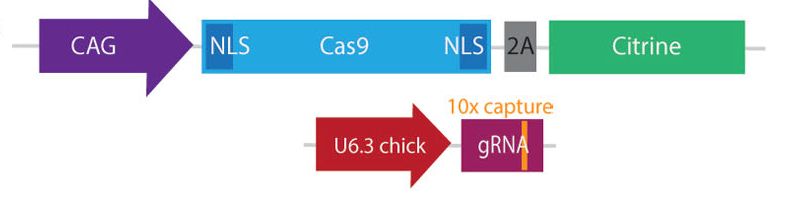
Key refinement: incorporating capture sequences into gRNAs to enable single-cell RNA-seq tracking of perturbations

