
Find out more: royalsocietypublishing.org/doi/10.1098/... #evolution #palaeontology #taxonomyandsystematics

Find out more: royalsocietypublishing.org/doi/10.1098/... #evolution #palaeontology #taxonomyandsystematics


Contact us if you would like to participate or to know more about it!
Among our 29 themed symposia + 1 open symposium, we are pleased to feature:
✨ Life in the Phanerozoic Oceans: Evolution, diversity and ecology in deep time marine ecosystems✨
📩 To participate in this symposium or get more information, contact the conveners: 👇

Contact us if you would like to participate or to know more about it!
(I would still love to see a densitree plot though!)
www.biorxiv.org/content/10.1...

(I would still love to see a densitree plot though!)
www.biorxiv.org/content/10.1...
So incredible to watch our undergrad students from the lab shine in front of such a knowledgeable audience at @cpeg-cpb25.bsky.social. They absolutely crushed it and I couldn’t be prouder!
@fernandalandim.bsky.social, Anna Clara Annes, Jorge Silva and Gabriela Karam.




So incredible to watch our undergrad students from the lab shine in front of such a knowledgeable audience at @cpeg-cpb25.bsky.social. They absolutely crushed it and I couldn’t be prouder!
@fernandalandim.bsky.social, Anna Clara Annes, Jorge Silva and Gabriela Karam.
We are putting together an amazing workshop with details to come - can't wait to share it with everyone!🥳

We are putting together an amazing workshop with details to come - can't wait to share it with everyone!🥳
🔗 academic.oup.com/mbe/article/42/6/msaf123/8170119
#evobio #molbio
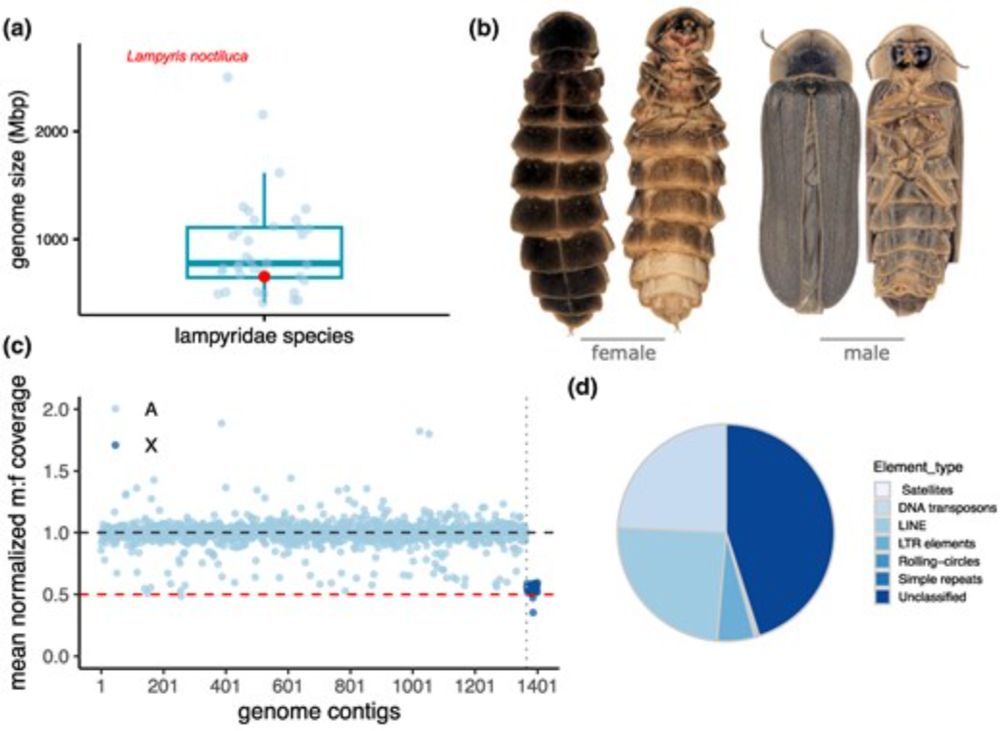


www.nature.com/articles/d41...

www.nature.com/articles/d41...
Ad here: jobs.uni-rostock.de/jobposting/c...

Ad here: jobs.uni-rostock.de/jobposting/c...
go.nature.com/4lFx4KN

go.nature.com/4lFx4KN
We introduce a covarion model for phylogenetic inference using discrete morphological data, addressing lineage- and character-specific rate heterogeneity.
www.biorxiv.org/content/10.1...
#phylogenetics #morphology #covarion #evolution

We introduce a covarion model for phylogenetic inference using discrete morphological data, addressing lineage- and character-specific rate heterogeneity.
www.biorxiv.org/content/10.1...
#phylogenetics #morphology #covarion #evolution
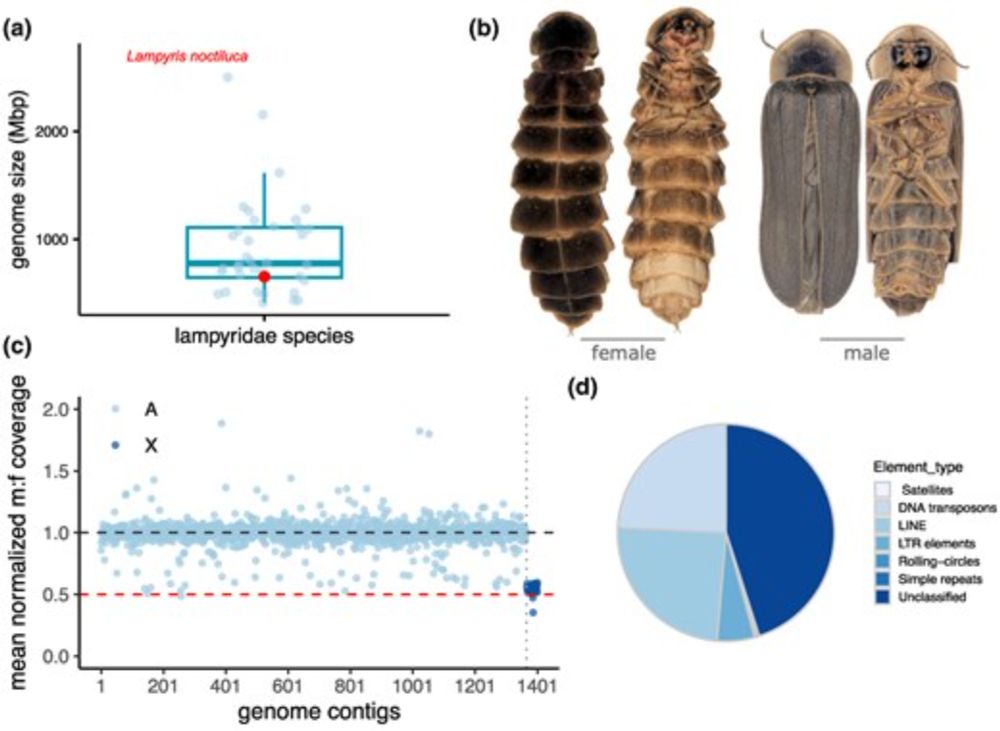
Model. Preprint coming soon!

Model. Preprint coming soon!
🔗 academic.oup.com/mbe/article/42/6/msaf123/8170119
#evobio #molbio

🔗 academic.oup.com/mbe/article/42/6/msaf123/8170119
#evobio #molbio

📍 Museum für Naturkunde, Berlin, Germany
🕒 3 Years, fully funded by the Leibniz Junior Researchgroup program
Details and Application portal 👇
jobs.museumfuernaturkunde.berlin/jobposting/8...
📍 Museum für Naturkunde, Berlin, Germany
🕒 3 Years, fully funded by the Leibniz Junior Researchgroup program
Details and Application portal 👇
jobs.museumfuernaturkunde.berlin/jobposting/8...
A new Marie Skłodowska-Curie Actions Postdoctoral Fellowships 2025 call is now open.
With a budget of €404.3 million, it will support around 1,650 researchers from Europe and beyond.
Apply by 10 September → europa.eu/!fBTMgF

A new Marie Skłodowska-Curie Actions Postdoctoral Fellowships 2025 call is now open.
With a budget of €404.3 million, it will support around 1,650 researchers from Europe and beyond.
Apply by 10 September → europa.eu/!fBTMgF
marie-sklodowska-curie-actions.ec.europa.eu/news/msca-op...

marie-sklodowska-curie-actions.ec.europa.eu/news/msca-op...


