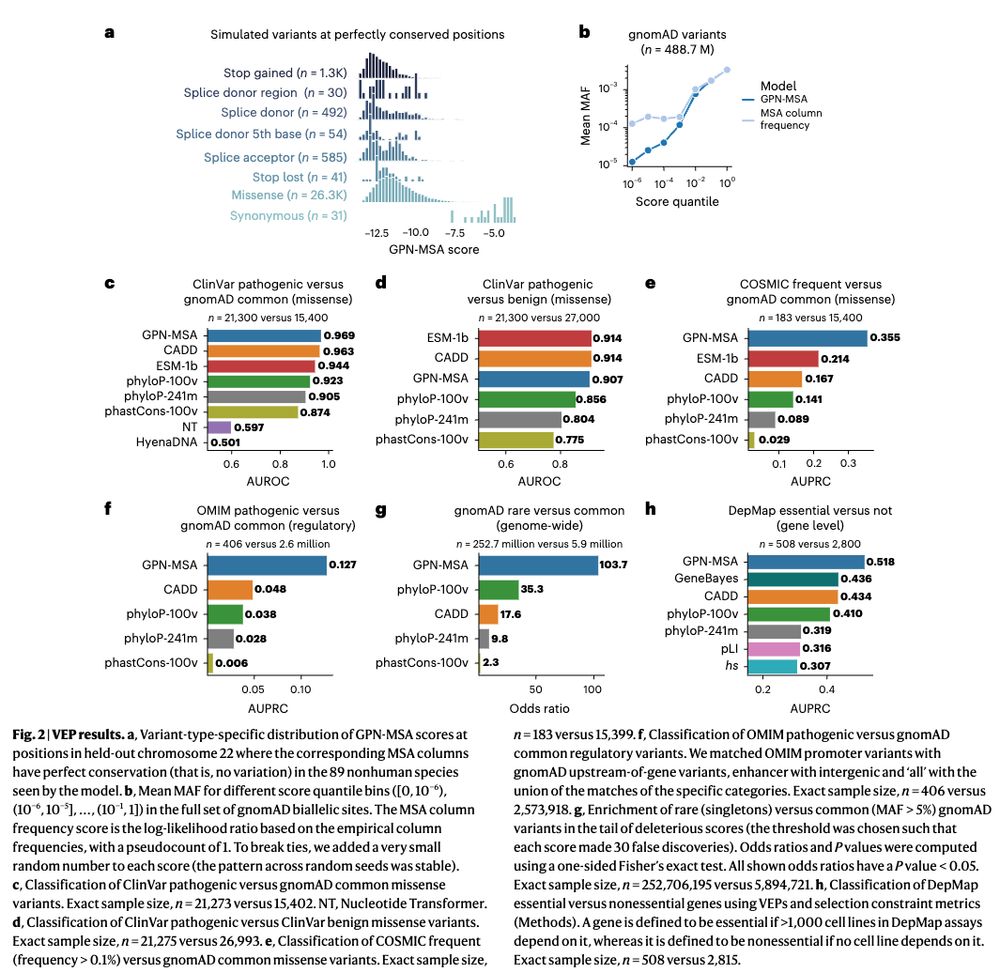
Climente-Gonzalez, Oh et al. develop an interpretable #ML model that integrates plasma proteomics with clinical risk factors to improve risk prediction for CVD 🫀 .
https://bit.ly/3ZUqyY8
@hclimente.eu @nkoell.bsky.social
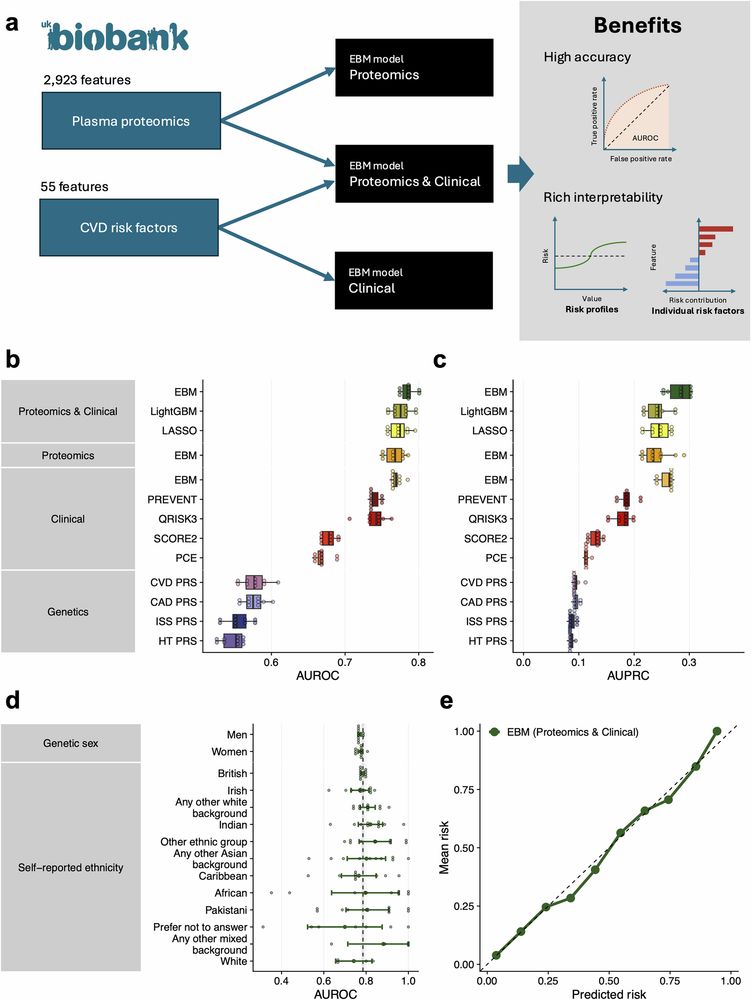
Climente-Gonzalez, Oh et al. develop an interpretable #ML model that integrates plasma proteomics with clinical risk factors to improve risk prediction for CVD 🫀 .
https://bit.ly/3ZUqyY8
@hclimente.eu @nkoell.bsky.social
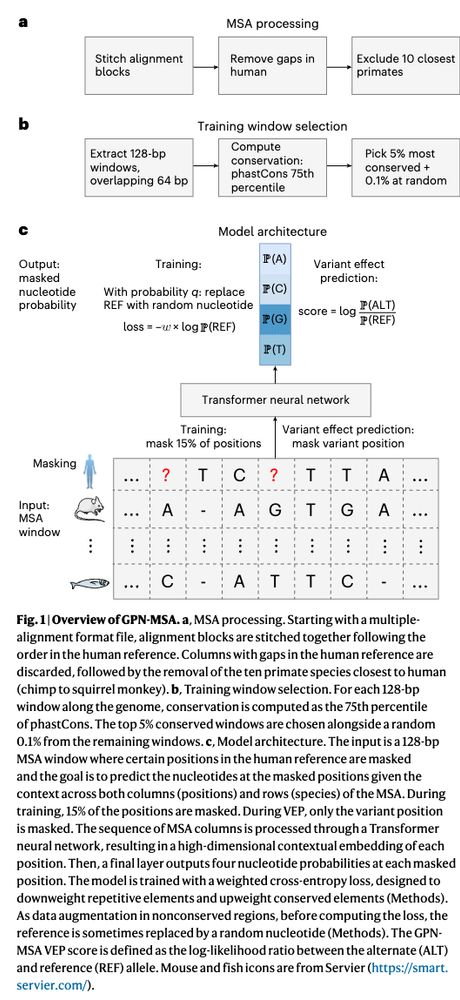
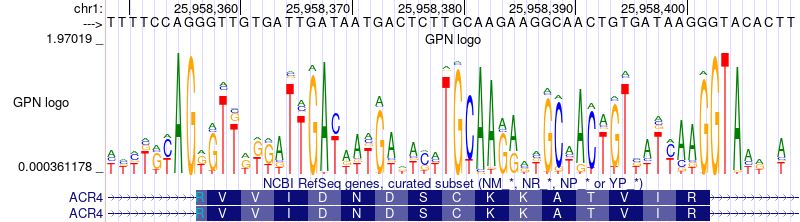
If you're keen on seeing how LLM libraries can be applied to DNA, my latest post breaks it all down. Give it a read: hclimente.eu/blog/hf-tran...
If you're keen on seeing how LLM libraries can be applied to DNA, my latest post breaks it all down. Give it a read: hclimente.eu/blog/hf-tran...
Here's a short tutorial if you're curious: hclimente.eu/blog/python-...
Here's a short tutorial if you're curious: hclimente.eu/blog/python-...
#MachineLearning #ExplainableAI #XAI #SHAP
#MachineLearning #ExplainableAI #XAI #SHAP

doi.org/10.1101/2025...
1/n
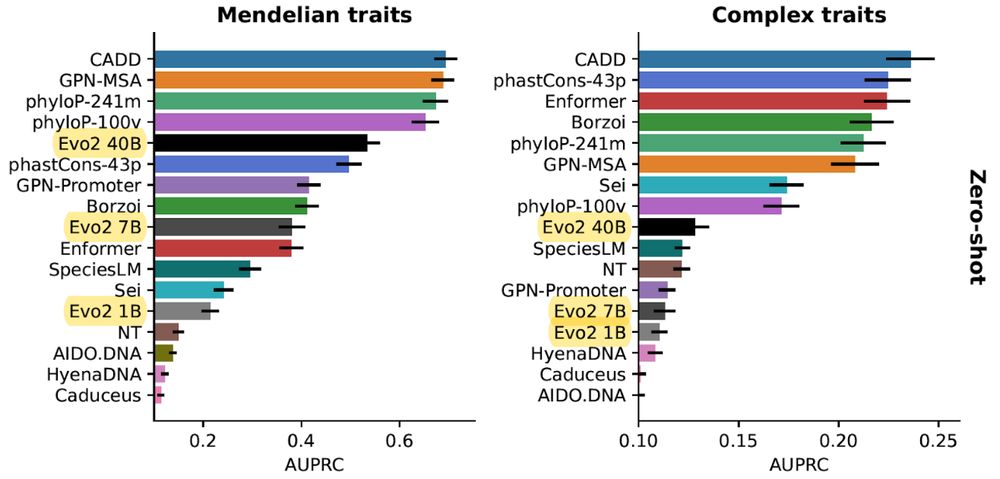
doi.org/10.1101/2025...
1/n